You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000282_01601
You are here: Home > Sequence: MGYG000000282_01601
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | TF01-11 sp003524945 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; TF01-11; TF01-11 sp003524945 | |||||||||||
| CAZyme ID | MGYG000000282_01601 | |||||||||||
| CAZy Family | GH59 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 85164; End: 88637 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH59 | 56 | 787 | 6.1e-172 | 0.9920760697305864 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02057 | Glyco_hydro_59 | 3.90e-96 | 62 | 394 | 1 | 292 | Glycosyl hydrolase family 59. |
| sd00036 | LRR_3 | 3.97e-11 | 1066 | 1137 | 25 | 95 | leucine-rich repeats. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
| sd00036 | LRR_3 | 4.29e-11 | 1066 | 1137 | 2 | 72 | leucine-rich repeats. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
| sd00036 | LRR_3 | 2.01e-10 | 1066 | 1120 | 71 | 124 | leucine-rich repeats. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
| pfam13306 | LRR_5 | 8.75e-10 | 1067 | 1137 | 1 | 68 | Leucine rich repeats (6 copies). This family includes a number of leucine rich repeats. This family contains a large number of BSPA-like surface antigens from Trichomonas vaginalis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNK59157.1 | 1.32e-290 | 45 | 925 | 315 | 1189 |
| ACL75594.1 | 2.54e-289 | 12 | 934 | 5 | 918 |
| ABG76970.1 | 2.54e-289 | 12 | 934 | 5 | 918 |
| AJQ96197.1 | 7.45e-288 | 44 | 925 | 308 | 1184 |
| QUL53185.1 | 2.21e-287 | 32 | 926 | 307 | 1193 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4CCC_A | 1.98e-20 | 59 | 787 | 22 | 650 | StructureOf Mouse Galactocerebrosidase With 4nbdg: Enzyme-substrate Complex [Mus musculus],4CCD_A Structure Of Mouse Galactocerebrosidase With D-galactal: Enzyme-intermediate Complex [Mus musculus],4CCE_A Structure Of Mouse Galactocerebrosidase With Galactose: Enzyme-product Complex [Mus musculus],4UFH_A Mouse Galactocerebrosidase complexed with iso-galacto-fagomine IGF [Mus musculus],4UFI_A Mouse Galactocerebrosidase complexed with aza-galacto-fagomine AGF [Mus musculus],4UFJ_A Mouse Galactocerebrosidase complexed with iso-galacto-fagomine lactam IGL [Mus musculus],4UFK_A Mouse Galactocerebrosidase complexed with dideoxy-imino-lyxitol DIL [Mus musculus],4UFL_A Mouse Galactocerebrosidase complexed with deoxy-galacto-noeurostegine DGN [Mus musculus],4UFM_A Mouse Galactocerebrosidase complexed with 1-deoxy-galacto-nojirimycin DGJ [Mus musculus],5NXB_A Mouse galactocerebrosidase in complex with saposin A [Mus musculus],5NXB_B Mouse galactocerebrosidase in complex with saposin A [Mus musculus],6Y6S_A Chain A, Galactocerebrosidase [Mus musculus],6Y6T_A Chain A, Galactocerebrosidase [Mus musculus] |
| 3ZR5_A | 1.99e-20 | 59 | 787 | 24 | 652 | STRUCTUREOF GALACTOCEREBROSIDASE FROM MOUSE [Mus musculus],3ZR6_A STRUCTURE OF GALACTOCEREBROSIDASE FROM MOUSE IN COMPLEX WITH GALACTOSE [Mus musculus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O02791 | 6.60e-23 | 12 | 787 | 5 | 681 | Galactocerebrosidase OS=Macaca mulatta OX=9544 GN=GALC PE=1 SV=2 |
| B5X3C1 | 1.89e-22 | 59 | 787 | 34 | 662 | Galactocerebrosidase OS=Salmo salar OX=8030 GN=galc PE=2 SV=1 |
| P54803 | 1.06e-21 | 12 | 787 | 5 | 681 | Galactocerebrosidase OS=Homo sapiens OX=9606 GN=GALC PE=1 SV=3 |
| P54804 | 3.07e-21 | 36 | 787 | 11 | 665 | Galactocerebrosidase OS=Canis lupus familiaris OX=9615 GN=GALC PE=1 SV=1 |
| P54818 | 9.72e-21 | 21 | 787 | 14 | 680 | Galactocerebrosidase OS=Mus musculus OX=10090 GN=Galc PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
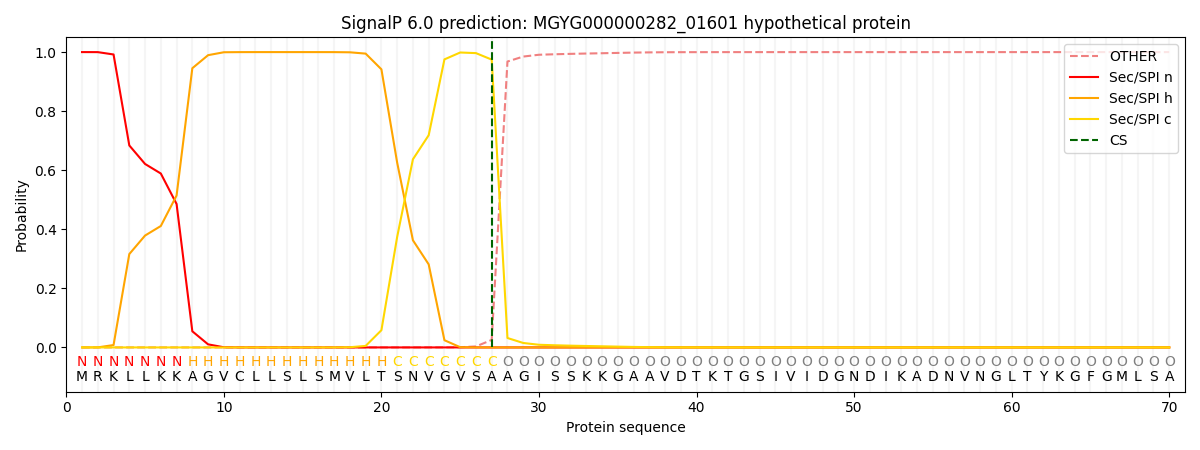
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000265 | 0.998989 | 0.000185 | 0.000189 | 0.000181 | 0.000157 |
