You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000282_01823
You are here: Home > Sequence: MGYG000000282_01823
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | TF01-11 sp003524945 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; TF01-11; TF01-11 sp003524945 | |||||||||||
| CAZyme ID | MGYG000000282_01823 | |||||||||||
| CAZy Family | GH59 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 25183; End: 28665 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH59 | 67 | 789 | 2.7e-173 | 0.9904912836767037 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02057 | Glyco_hydro_59 | 3.44e-99 | 72 | 404 | 1 | 292 | Glycosyl hydrolase family 59. |
| PRK11633 | PRK11633 | 5.06e-07 | 946 | 1039 | 51 | 148 | cell division protein DedD; Provisional |
| NF033761 | gliding_GltJ | 6.43e-07 | 948 | 1018 | 404 | 474 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| NF033761 | gliding_GltJ | 2.33e-06 | 944 | 1037 | 394 | 487 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| pfam15324 | TALPID3 | 3.09e-06 | 942 | 1036 | 924 | 1004 | Hedgehog signalling target. TALPID3 is a family of eukaryotic proteins that are targets for Hedgehog signalling. Mutations in this gene noticed first in chickens lead to multiple abnormalities of development. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ACL75594.1 | 1.76e-313 | 48 | 932 | 27 | 917 |
| ABG76970.1 | 1.76e-313 | 48 | 932 | 27 | 917 |
| AJQ96197.1 | 1.57e-308 | 54 | 924 | 308 | 1184 |
| QUL53185.1 | 1.18e-303 | 55 | 925 | 317 | 1193 |
| QNK59157.1 | 5.17e-303 | 37 | 924 | 298 | 1189 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q5SNX7 | 2.27e-25 | 69 | 790 | 30 | 658 | Galactocerebrosidase OS=Danio rerio OX=7955 GN=galc PE=2 SV=1 |
| Q498K0 | 1.49e-22 | 69 | 789 | 44 | 671 | Galactocerebrosidase OS=Xenopus laevis OX=8355 GN=galc PE=2 SV=2 |
| Q0VA39 | 5.99e-22 | 69 | 789 | 44 | 672 | Galactocerebrosidase OS=Xenopus tropicalis OX=8364 GN=galc PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
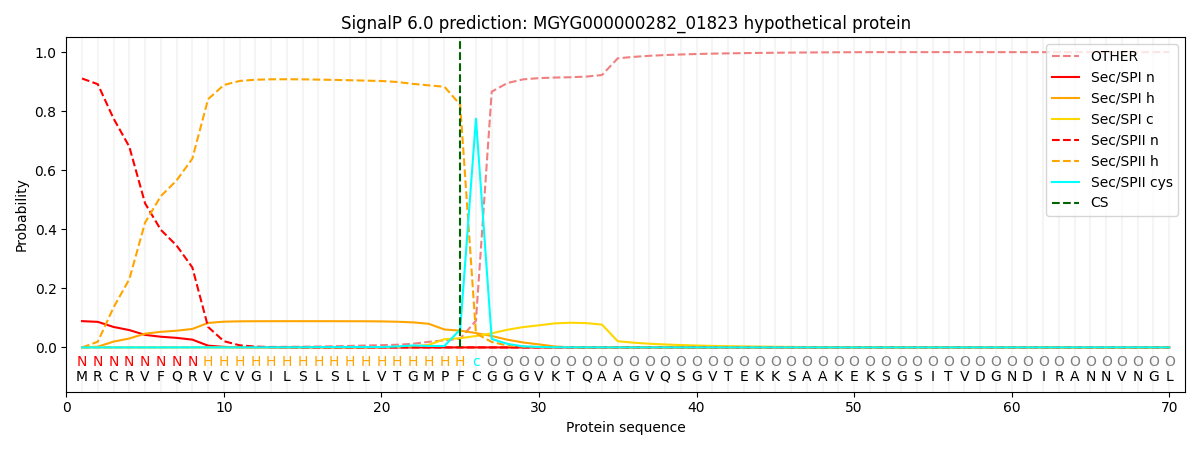
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000422 | 0.087435 | 0.911900 | 0.000149 | 0.000058 | 0.000031 |
