You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000282_02256
You are here: Home > Sequence: MGYG000000282_02256
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | TF01-11 sp003524945 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; TF01-11; TF01-11 sp003524945 | |||||||||||
| CAZyme ID | MGYG000000282_02256 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 39558; End: 41819 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 3.38e-18 | 404 | 635 | 45 | 244 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam08239 | SH3_3 | 2.21e-04 | 43 | 104 | 1 | 54 | Bacterial SH3 domain. |
| cd11765 | SH3_Nck_1 | 0.004 | 54 | 99 | 5 | 47 | First Src Homology 3 domain of Nck adaptor proteins. Nck adaptor proteins regulate actin cytoskeleton dynamics by linking proline-rich effector molecules to protein tyrosine kinases and phosphorylated signaling intermediates. They contain three SH3 domains and a C-terminal SH2 domain. They function downstream of the PDGFbeta receptor and are involved in Rho GTPase signaling and actin dynamics. Vertebrates contain two Nck adaptor proteins: Nck1 (also called Nckalpha) and Nck2 (also called Nckbeta or Growth factor receptor-bound protein 4, Grb4), which show partly overlapping functions but also bind distinct targets. Their SH3 domains are involved in recruiting downstream effector molecules, such as the N-WASP/Arp2/3 complex, which when activated induces actin polymerization that results in the production of pedestals, or protrusions of the plasma membrane. The first SH3 domain of Nck proteins preferentially binds the PxxDY sequence, which is present in the CD3e cytoplasmic tail. This binding inhibits phosphorylation by Src kinases, resulting in the downregulation of TCR surface expression. SH3 domains are protein interaction domains that usually bind to proline-rich ligands with moderate affinity and selectivity, preferentially a PxxP motif. They play versatile and diverse roles in the cell including the regulation of enzymes, changing the subcellular localization of signaling pathway components, and mediating the formation of multiprotein complex assemblies. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHQ61172.1 | 1.85e-153 | 33 | 745 | 33 | 760 |
| BCN29841.1 | 1.31e-152 | 1 | 751 | 1 | 888 |
| BCJ97942.1 | 6.51e-144 | 36 | 751 | 152 | 906 |
| BCJ93456.1 | 5.06e-142 | 2 | 751 | 4 | 894 |
| CUH91961.1 | 2.37e-135 | 41 | 751 | 145 | 911 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4PI7_A | 1.35e-08 | 469 | 626 | 76 | 222 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PI9_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50],4PIA_A Chain A, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 4PI8_A | 8.08e-08 | 469 | 626 | 76 | 222 | ChainA, Autolysin E [Staphylococcus aureus subsp. aureus Mu50] |
| 6FXO_A | 1.75e-07 | 407 | 635 | 37 | 243 | ChainA, Bifunctional autolysin [Staphylococcus aureus subsp. aureus Mu50] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39848 | 2.61e-10 | 407 | 635 | 679 | 879 | Beta-N-acetylglucosaminidase OS=Bacillus subtilis (strain 168) OX=224308 GN=lytD PE=1 SV=1 |
| Q8CPQ1 | 3.10e-08 | 407 | 635 | 1128 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=atl PE=3 SV=1 |
| O33635 | 7.02e-08 | 407 | 635 | 1128 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis OX=1282 GN=atl PE=1 SV=1 |
| Q5HQB9 | 7.02e-08 | 407 | 635 | 1128 | 1334 | Bifunctional autolysin OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=atl PE=3 SV=1 |
| Q6GAG0 | 3.13e-06 | 407 | 635 | 1043 | 1249 | Bifunctional autolysin OS=Staphylococcus aureus (strain MSSA476) OX=282459 GN=atl PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
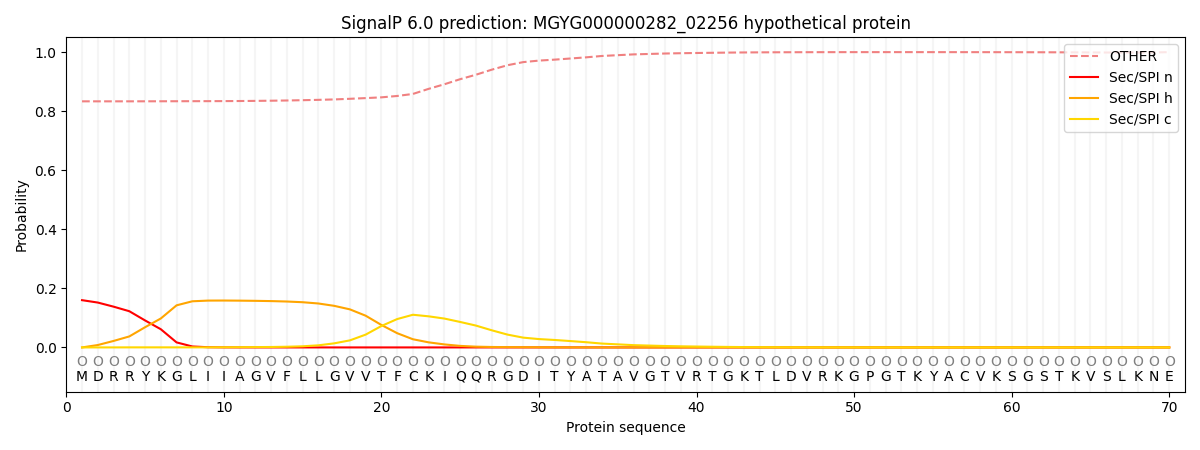
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.841010 | 0.150809 | 0.007172 | 0.000350 | 0.000216 | 0.000446 |

