You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000282_02820
You are here: Home > Sequence: MGYG000000282_02820
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | TF01-11 sp003524945 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; TF01-11; TF01-11 sp003524945 | |||||||||||
| CAZyme ID | MGYG000000282_02820 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 24034; End: 27948 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 348 | 630 | 2.3e-74 | 0.9927536231884058 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 4.25e-44 | 348 | 631 | 15 | 270 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.14e-18 | 312 | 593 | 34 | 327 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| NF033761 | gliding_GltJ | 2.30e-07 | 1138 | 1212 | 410 | 481 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
| PRK14951 | PRK14951 | 2.87e-07 | 1131 | 1208 | 375 | 443 | DNA polymerase III subunits gamma and tau; Provisional |
| NF033761 | gliding_GltJ | 4.15e-07 | 1137 | 1207 | 421 | 490 | adventurous gliding motility protein GltJ. Adventurous gliding motility protein GltJ, also known as AgmX, occurs in delta-proteobacteria such as Myxococcus xanthus. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AUO18792.1 | 4.49e-248 | 201 | 1131 | 136 | 1046 |
| AUO19859.1 | 3.58e-163 | 241 | 856 | 33 | 642 |
| BCN29385.1 | 4.16e-56 | 317 | 819 | 349 | 844 |
| AIQ47948.1 | 1.06e-55 | 317 | 645 | 36 | 394 |
| AIQ53446.1 | 4.89e-55 | 317 | 645 | 36 | 394 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2JEP_A | 6.67e-52 | 321 | 666 | 34 | 393 | Nativefamily 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEP_B Native family 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEQ_A Family 5 xyloglucanase from Paenibacillus pabuli in complex with ligand [Paenibacillus pabuli] |
| 6WQP_A | 5.45e-46 | 314 | 592 | 8 | 294 | GH5-4broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQP_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQV_A GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_C GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_D GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis] |
| 6Q1I_A | 5.88e-46 | 324 | 666 | 16 | 352 | GH5-4broad specificity endoglucanase from Clostrdium longisporum [Clostridium longisporum],6Q1I_B GH5-4 broad specificity endoglucanase from Clostrdium longisporum [Clostridium longisporum] |
| 6PZ7_A | 9.68e-45 | 320 | 630 | 7 | 305 | GH5-4broad specificity endoglucanase from Clostridium acetobutylicum [Clostridium acetobutylicum ATCC 824] |
| 5D9N_A | 2.37e-43 | 326 | 592 | 17 | 286 | Crystalstructure of PbGH5A, a glycoside hydrolase family 5 member from Prevotella bryantii B14, in complex with the xyloglucan heptasaccharide XXXG [Prevotella bryantii],5D9N_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 member from Prevotella bryantii B14, in complex with the xyloglucan heptasaccharide XXXG [Prevotella bryantii],5D9P_A Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, in complex with an inhibitory N-bromoacetylglycosylamine derivative of XXXG [Prevotella bryantii],5D9P_B Crystal structure of PbGH5A, a glycoside hydrolase family 5 enzyme from Prevotella bryantii B14, in complex with an inhibitory N-bromoacetylglycosylamine derivative of XXXG [Prevotella bryantii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O08342 | 3.36e-48 | 305 | 666 | 23 | 398 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
| P54937 | 1.67e-44 | 324 | 666 | 41 | 377 | Endoglucanase A OS=Clostridium longisporum OX=1523 GN=celA PE=1 SV=1 |
| P23660 | 1.09e-43 | 324 | 663 | 29 | 359 | Endoglucanase A OS=Ruminococcus albus OX=1264 GN=celA PE=1 SV=1 |
| P28621 | 5.04e-42 | 318 | 692 | 38 | 397 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
| P17901 | 6.66e-40 | 326 | 665 | 51 | 398 | Endoglucanase A OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
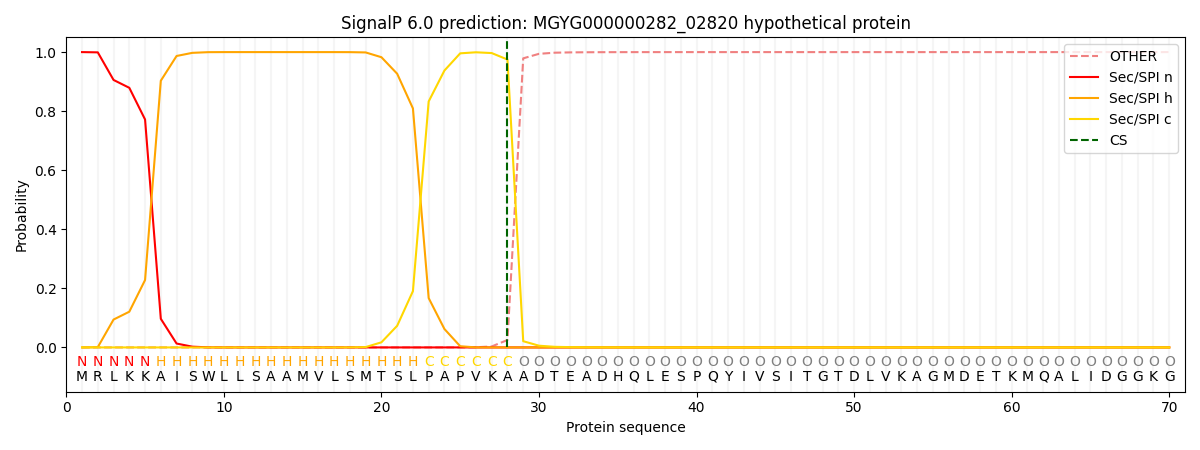
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000204 | 0.999143 | 0.000172 | 0.000179 | 0.000156 | 0.000142 |
