You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000284_00103
You are here: Home > Sequence: MGYG000000284_00103
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
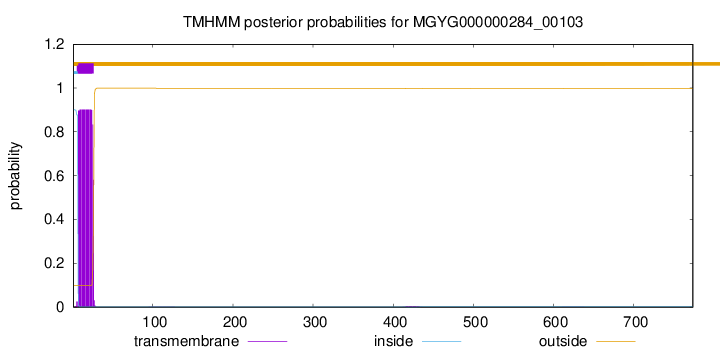
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp900540715 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp900540715 | |||||||||||
| CAZyme ID | MGYG000000284_00103 | |||||||||||
| CAZy Family | GH95 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 156215; End: 158539 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 135 | 732 | 3.9e-66 | 0.8296398891966759 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1554 | ATH1 | 1.34e-04 | 110 | 542 | 127 | 548 | Trehalose and maltose hydrolase (possible phosphorylase) [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHW32096.1 | 1.44e-107 | 122 | 769 | 428 | 1081 |
| QXE33659.1 | 2.43e-97 | 130 | 769 | 306 | 952 |
| AWS40658.1 | 5.63e-97 | 155 | 772 | 305 | 929 |
| BCB76148.1 | 1.21e-89 | 135 | 769 | 237 | 879 |
| QKW17790.1 | 5.67e-82 | 21 | 767 | 35 | 761 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7KMQ_A | 4.46e-08 | 302 | 730 | 335 | 757 | ChainA, Glyco_hyd_65N_2 domain-containing protein [Xanthomonas citri pv. citri str. 306],7KMQ_B Chain B, Glyco_hyd_65N_2 domain-containing protein [Xanthomonas citri pv. citri str. 306] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8L7W8 | 6.09e-10 | 306 | 724 | 379 | 806 | Alpha-L-fucosidase 2 OS=Arabidopsis thaliana OX=3702 GN=FUC95A PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
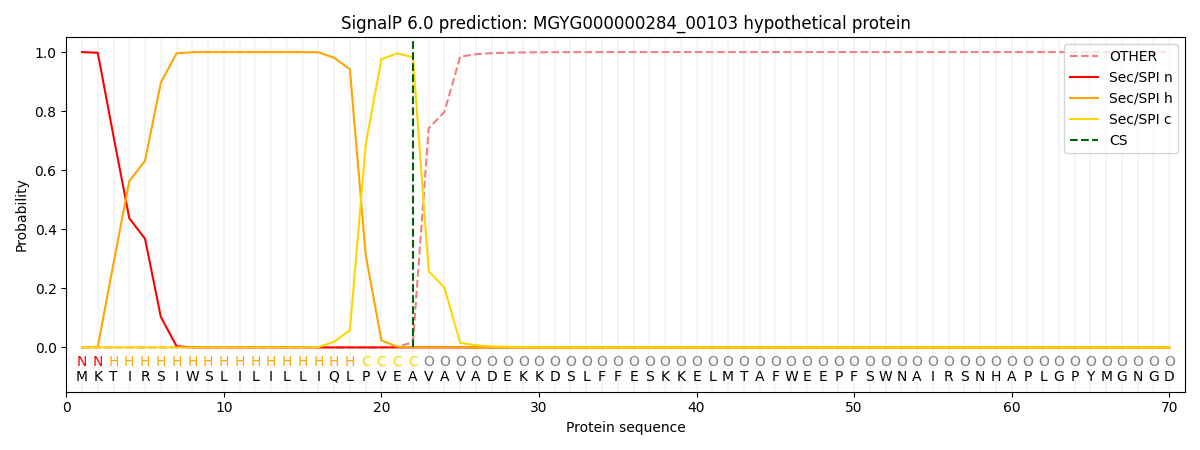
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000235 | 0.999178 | 0.000155 | 0.000142 | 0.000129 | 0.000130 |