You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000284_00118
You are here: Home > Sequence: MGYG000000284_00118
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp900540715 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp900540715 | |||||||||||
| CAZyme ID | MGYG000000284_00118 | |||||||||||
| CAZy Family | CBM6 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 188687; End: 190969 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH120 | 450 | 539 | 1.8e-31 | 0.978021978021978 |
| CBM6 | 223 | 306 | 3.6e-17 | 0.5797101449275363 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd04084 | CBM6_xylanase-like | 1.87e-19 | 223 | 309 | 27 | 112 | Carbohydrate Binding Module 6 (CBM6); many are appended to glycoside hydrolase (GH) family 11 and GH43 xylanase domains. This family includes carbohydrate binding module 6 (CBM6) domains that are appended mainly to glycoside hydrolase (GH) family domains, including GH3, GH11, and GH43 domains. These CBM6s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. Examples of proteins having CMB6s belonging to this family are Microbispora bispora GghA, a 1,4-beta-D-glucan glucohydrolase (GH3); Clostridium thermocellum xylanase U (GH11), and Penicillium purpurogenum ABF3, a bifunctional alpha-L-arabinofuranosidase/xylobiohydrolase (GH43). GH3 comprises enzymes with activities including beta-glucosidase (hydrolyzes beta-galactosidase) and beta-xylosidase (hydrolyzes 1,4-beta-D-xylosidase). GH11 family comprises enzymes with xylanase (endo-1,4-beta-xylanase) activity which catalyze the hydrolysis of beta-1,4 bonds of xylan, the major component of hemicelluloses, to generate xylooligosaccharides and xylose. GH43 includes beta-xylosidases and beta-xylanases, using aryl-glycosides as substrates. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. |
| smart00606 | CBD_IV | 9.60e-11 | 223 | 310 | 34 | 121 | Cellulose Binding Domain Type IV. |
| pfam03422 | CBM_6 | 2.26e-10 | 223 | 307 | 26 | 110 | Carbohydrate binding module (family 6). |
| pfam13229 | Beta_helix | 3.35e-10 | 445 | 578 | 29 | 155 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 8.09e-09 | 386 | 540 | 35 | 138 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AOW10547.1 | 0.0 | 10 | 758 | 9 | 761 |
| AOW08578.1 | 0.0 | 19 | 758 | 18 | 757 |
| AOW10546.1 | 0.0 | 10 | 758 | 9 | 760 |
| ARD26815.1 | 2.84e-204 | 8 | 756 | 12 | 757 |
| ARN56258.1 | 3.59e-199 | 7 | 755 | 13 | 761 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3VST_A | 1.90e-130 | 21 | 756 | 2 | 638 | Thecomplex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VST_B The complex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VST_C The complex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VST_D The complex structure of XylC with Tris [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_A The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_B The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_C The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSU_D The complex structure of XylC with xylobiose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_A The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_B The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_C The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485],3VSV_D The complex structure of XylC with xylose [Thermoanaerobacterium saccharolyticum JW/SL-YS485] |
| 1GMM_A | 1.37e-07 | 223 | 308 | 33 | 118 | ChainA, CBM6 [Acetivibrio thermocellus],1UXX_X Chain X, Xylanase U [Acetivibrio thermocellus] |
| 1UY1_A | 2.40e-07 | 218 | 308 | 45 | 133 | Bindingsub-site dissection of a family 6 carbohydrate-binding module by X-ray crystallography and isothermal titration calorimetry [Thermoclostridium stercorarium],1UY2_A Binding sub-site dissection of a family 6 carbohydrate-binding module by X-ray crystallography and isothermal titration calorimetry [Thermoclostridium stercorarium],1UY3_A Binding sub-site dissection of a family 6 carbohydrate-binding module by X-ray crystallography and isothermal titration calorimetry [Thermoclostridium stercorarium],1UY4_A Binding sub-site dissection of a family 6 carbohydrate-binding module by X-ray crystallography and isothermal titration calorimetry [Thermoclostridium stercorarium] |
| 1W9S_A | 1.94e-06 | 223 | 284 | 41 | 102 | ChainA, Bh0236 Protein [Halalkalibacterium halodurans],1W9S_B Chain B, Bh0236 Protein [Halalkalibacterium halodurans],1W9T_A Chain A, Bh0236 Protein [Halalkalibacterium halodurans],1W9T_B Chain B, Bh0236 Protein [Halalkalibacterium halodurans],1W9W_A Chain A, Bh0236 Protein [Halalkalibacterium halodurans] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
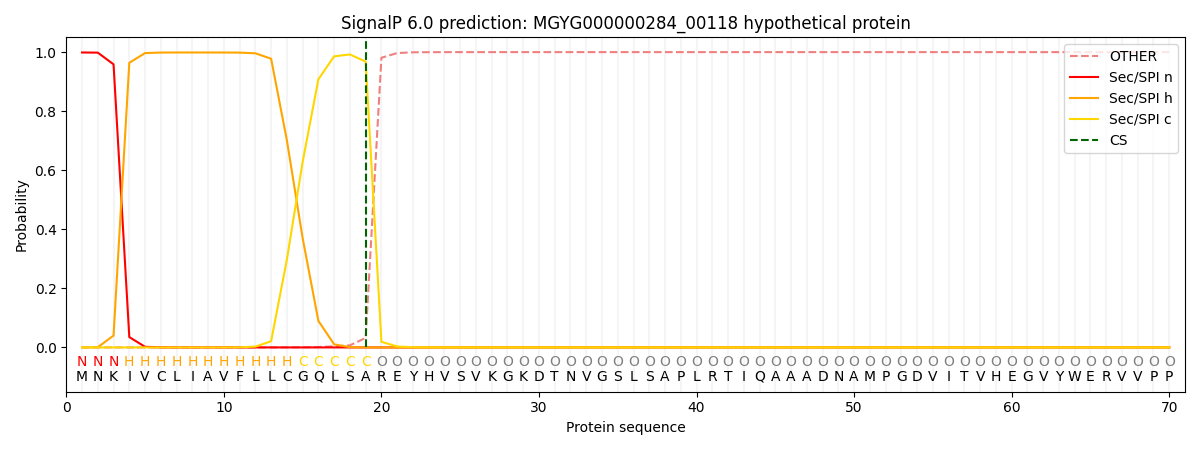
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000521 | 0.995994 | 0.002818 | 0.000223 | 0.000221 | 0.000207 |
