You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000284_00241
You are here: Home > Sequence: MGYG000000284_00241
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp900540715 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp900540715 | |||||||||||
| CAZyme ID | MGYG000000284_00241 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 98592; End: 102206 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 36 | 747 | 1.8e-87 | 0.8264563106796117 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 2.02e-26 | 196 | 688 | 354 | 870 | alpha-L-rhamnosidase. |
| pfam02837 | Glyco_hydro_2_N | 1.58e-06 | 1094 | 1178 | 56 | 142 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
| PRK10150 | PRK10150 | 5.51e-05 | 1091 | 1172 | 53 | 137 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 1.46e-04 | 1107 | 1178 | 113 | 186 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam08531 | Bac_rhamnosid_N | 8.47e-04 | 730 | 792 | 101 | 147 | Alpha-L-rhamnosidase N-terminal domain. This family consists of bacterial rhamnosidase A and B enzymes. This domain is probably involved in substrate recognition. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBL01784.1 | 0.0 | 1 | 1202 | 1 | 1202 |
| BBL12449.1 | 0.0 | 1 | 1202 | 1 | 1202 |
| BBL09655.1 | 0.0 | 1 | 1202 | 1 | 1202 |
| QGY44259.1 | 0.0 | 1 | 1198 | 4 | 1194 |
| QGQ95144.1 | 0.0 | 24 | 1200 | 29 | 1237 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQM_A | 3.45e-09 | 1091 | 1175 | 972 | 1059 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 3.45e-09 | 1091 | 1175 | 972 | 1059 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 6D89_A | 1.57e-07 | 1086 | 1171 | 81 | 168 | Bacteroidesuniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_B Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_C Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis],6D89_D Bacteroides uniformis beta-glucuronidase 1 with N-terminal loop deletion [Bacteroides uniformis] |
| 6D1N_A | 1.58e-07 | 1086 | 1171 | 89 | 176 | Apostructure of Bacteroides uniformis Beta-glucuronidase 1 [Bacteroides uniformis],6D1N_B Apo structure of Bacteroides uniformis Beta-glucuronidase 1 [Bacteroides uniformis],6D41_A Bacteriodes uniformis beta-glucuronidase 1 bound to D-glucaro-1,5-lactone [Bacteroides uniformis],6D41_B Bacteriodes uniformis beta-glucuronidase 1 bound to D-glucaro-1,5-lactone [Bacteroides uniformis],6D6W_A Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D6W_B Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D6W_C Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D6W_D Bacteroides uniformis beta-glucuronidase 1 bound to glucuronate [Bacteroides uniformis],6D7F_A Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_B Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_C Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_D Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_E Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis],6D7F_F Bacteroides uniformis beta-glucuronidase 1 bound to thiophenyl-beta-D-glucuronide [Bacteroides uniformis] |
| 7VQM_A | 3.22e-06 | 1090 | 1171 | 80 | 163 | ChainA, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_B Chain B, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_C Chain C, GH2 beta-galacturonate AqGalA [Aquimarina sp.],7VQM_D Chain D, GH2 beta-galacturonate AqGalA [Aquimarina sp.] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KNA8 | 5.61e-27 | 35 | 1175 | 29 | 1093 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22040 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
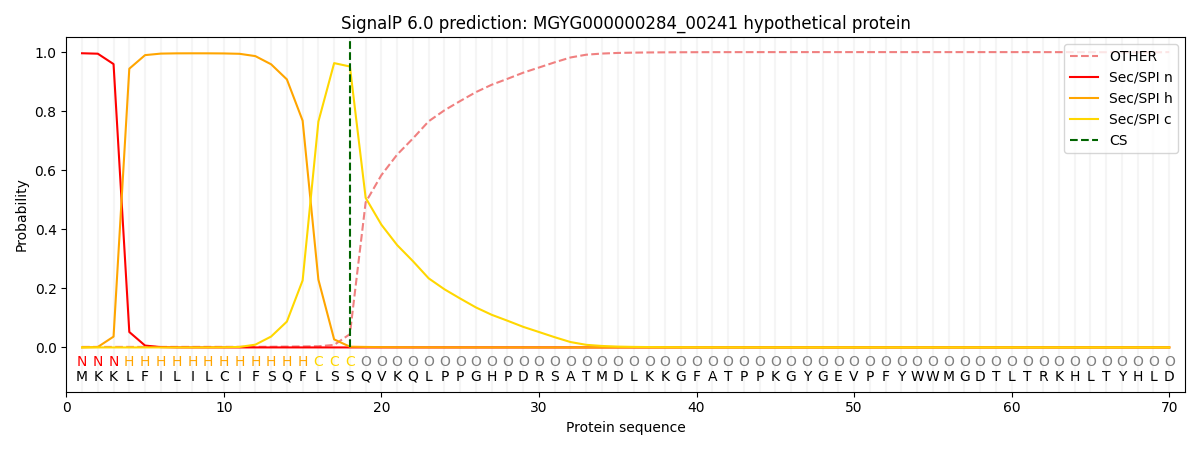
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.004506 | 0.991754 | 0.002998 | 0.000275 | 0.000232 | 0.000235 |
