You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000284_00763
You are here: Home > Sequence: MGYG000000284_00763
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp900540715 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp900540715 | |||||||||||
| CAZyme ID | MGYG000000284_00763 | |||||||||||
| CAZy Family | GH156 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 56858; End: 58510 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH156 | 25 | 545 | 7.1e-115 | 0.9402173913043478 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1649 | YddW | 7.46e-08 | 128 | 306 | 113 | 309 | Uncharacterized lipoprotein YddW, UPF0748 family [Function unknown]. |
| pfam02638 | GHL10 | 8.83e-05 | 118 | 310 | 58 | 267 | Glycosyl hydrolase-like 10. This is family of bacterial glycosyl-hydrolase-like proteins falling into the family GHL10 as described above,. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT49878.1 | 0.0 | 1 | 550 | 1 | 550 |
| BBD44721.1 | 1.54e-219 | 59 | 550 | 3 | 491 |
| BCG53795.1 | 8.78e-195 | 20 | 550 | 8 | 543 |
| QGA27036.1 | 7.58e-174 | 3 | 550 | 2 | 557 |
| QDU82332.1 | 5.68e-42 | 31 | 380 | 28 | 383 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6RZD_A | 1.92e-38 | 53 | 530 | 25 | 493 | Crystalstructure of an inverting family GH156 exosialidase from uncultured bacterium pG7 [uncultured bacterium pG7],6RZD_B Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 [uncultured bacterium pG7],6S00_A Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-acetylneuraminic acid [uncultured bacterium pG7],6S00_B Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-acetylneuraminic acid [uncultured bacterium pG7],6S0E_A Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-Acetyl-2,3-dehydro-2-deoxyneuraminic acid [uncultured bacterium pG7],6S0E_B Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-Acetyl-2,3-dehydro-2-deoxyneuraminic acid [uncultured bacterium pG7],6S0F_A Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with 3-Deoxy-D-glycero-D-galacto-2-nonulosonic acid [uncultured bacterium pG7],6S0F_B Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with 3-Deoxy-D-glycero-D-galacto-2-nonulosonic acid [uncultured bacterium pG7] |
| 6S04_A | 1.01e-33 | 53 | 530 | 25 | 493 | Crystalstructure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-glycolylneuraminic acid [uncultured bacterium pG7],6S04_B Crystal structure of an inverting family GH156 exosialidase from uncultured bacterium pG7 in complex with N-glycolylneuraminic acid [uncultured bacterium pG7] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
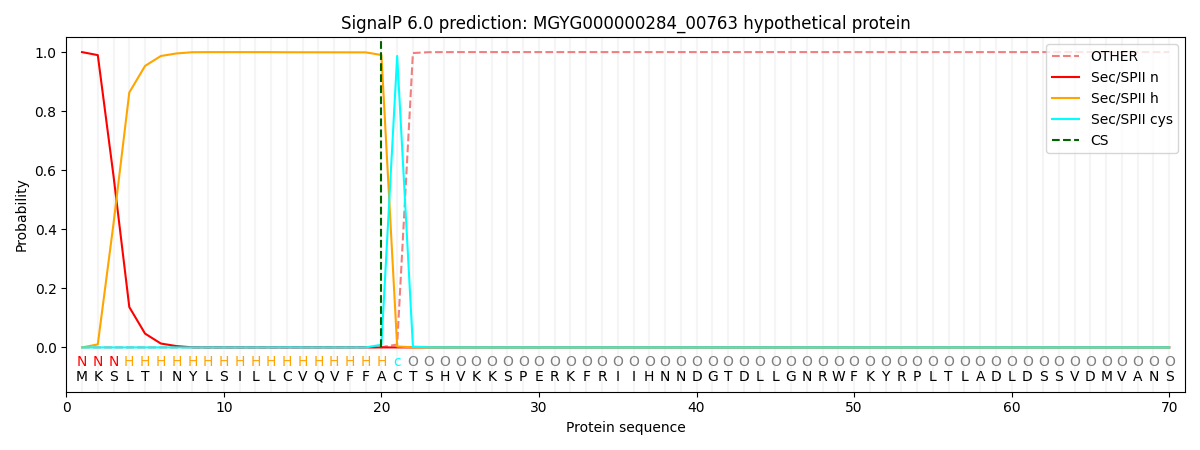
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000060 | 0.999989 | 0.000000 | 0.000000 | 0.000000 |
