You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000284_01599
You are here: Home > Sequence: MGYG000000284_01599
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Parabacteroides sp900540715 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp900540715 | |||||||||||
| CAZyme ID | MGYG000000284_01599 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 19739; End: 22618 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 28 | 732 | 6.7e-64 | 0.7207446808510638 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 2.31e-19 | 22 | 451 | 3 | 426 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10150 | PRK10150 | 4.40e-11 | 29 | 449 | 10 | 440 | beta-D-glucuronidase; Provisional |
| PRK10340 | ebgA | 1.10e-09 | 97 | 431 | 112 | 449 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam00703 | Glyco_hydro_2 | 3.98e-09 | 204 | 308 | 3 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| pfam02836 | Glyco_hydro_2_C | 6.85e-07 | 310 | 450 | 1 | 154 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT50289.1 | 0.0 | 1 | 957 | 1 | 956 |
| QIX67210.1 | 0.0 | 25 | 959 | 19 | 939 |
| BBK91423.1 | 0.0 | 25 | 959 | 10 | 930 |
| QUR50384.1 | 0.0 | 25 | 959 | 19 | 939 |
| QKH97806.1 | 0.0 | 25 | 959 | 19 | 939 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6D8K_A | 6.49e-13 | 33 | 456 | 37 | 442 | Bacteroidesmultiple species beta-glucuronidase [Bacteroides ovatus],6D8K_B Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_C Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_D Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus] |
| 6ETZ_A | 1.07e-11 | 31 | 491 | 36 | 482 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB [Arthrobacter sp. 32cB],6H1P_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB - data collected at room temperature [Arthrobacter sp. 32cB] |
| 6SEB_A | 1.07e-11 | 31 | 491 | 57 | 503 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB in complex with IPTG [Arthrobacter sp. 32cB],6SEC_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cBon complex with ONPG [Arthrobacter sp. 32cB],6SED_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB in complex with galactose [Arthrobacter sp. 32cB] |
| 6ZJP_A | 1.07e-11 | 31 | 491 | 57 | 503 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB mutant E517Q [Arthrobacter sp. 32cB] |
| 6ZJQ_A | 1.08e-11 | 31 | 491 | 59 | 505 | Cold-adaptedbeta-D-galactosidase from Arthrobacter sp. 32cB mutant E517Q in complex with galactose [Arthrobacter sp. 32cB],6ZJR_A Cold-adapted beta-D-galactosidase from Arthrobacter sp. 32cB mutant E517Q in complex with lactulose [Arthrobacter sp. 32cB] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23989 | 1.76e-10 | 27 | 550 | 41 | 572 | Beta-galactosidase OS=Streptococcus thermophilus OX=1308 GN=lacZ PE=3 SV=1 |
| T2KM09 | 8.12e-07 | 125 | 465 | 138 | 465 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22050 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
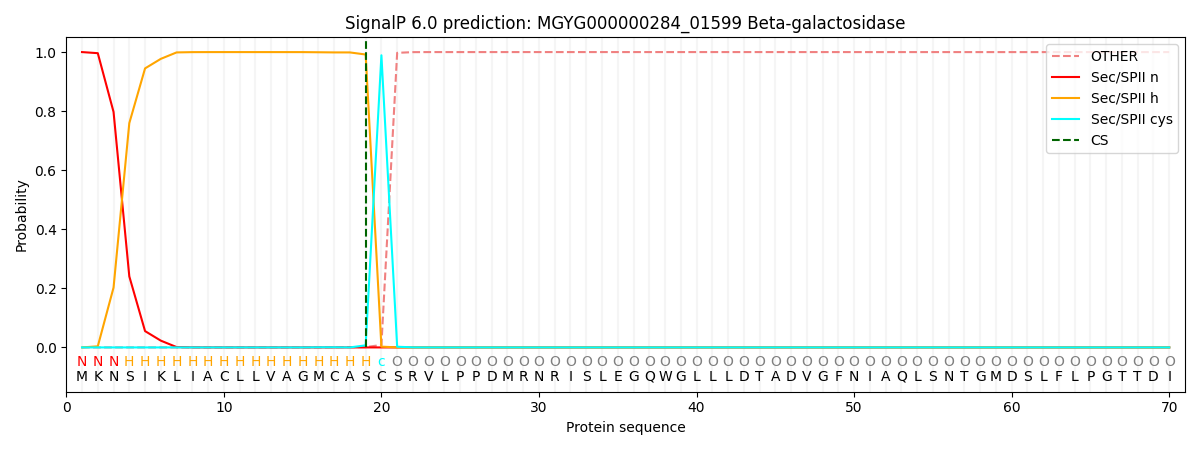
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000058 | 0.000000 | 0.000000 | 0.000000 |
