You are browsing environment: HUMAN GUT
MGYG000000284_04469
Basic Information
help
Species
Parabacteroides sp900540715
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides; Parabacteroides sp900540715
CAZyme ID
MGYG000000284_04469
CAZy Family
GT83
CAZyme Description
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase
CAZyme Property
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000000284
5935659
MAG
Sweden
Europe
Gene Location
Start: 7051;
End: 8619
Strand: -
No EC number prediction in MGYG000000284_04469.
Family
Start
End
Evalue
family coverage
GT83
15
429
6.7e-69
0.7759259259259259
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
COG1807
ArnT
1.62e-23
15
401
15
411
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis].
more
PRK13279
arnT
5.31e-09
12
323
5
322
lipid IV(A) 4-amino-4-deoxy-L-arabinosyltransferase.
more
pfam13231
PMT_2
2.23e-05
63
209
1
146
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366.
more
pfam03901
Glyco_transf_22
2.26e-04
82
357
78
354
Alg9-like mannosyltransferase family. Members of this family are mannosyltransferase enzymes. At least some members are localized in endoplasmic reticulum and involved in GPI anchor biosynthesis.
more
Hit ID
E-Value
Query Start
Query End
Hit Start
Hit End
Description
O67270
6.12e-22
34
341
29
328
Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1
more
This protein is predicted as OTHER
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.999982
0.000017
0.000001
0.000000
0.000000
0.000007
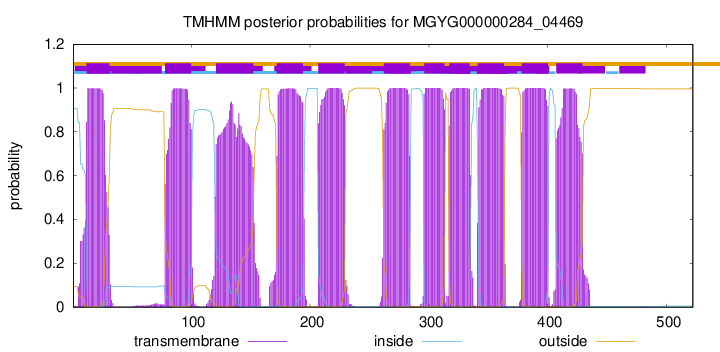
start
end
12
31
78
100
121
152
172
194
207
229
262
284
296
313
317
334
341
363
378
400
407
429