You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000285_00065
You are here: Home > Sequence: MGYG000000285_00065
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
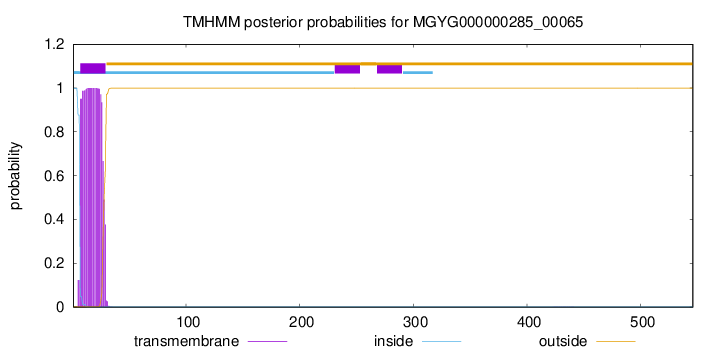
TMHMM annotations
Basic Information help
| Species | UBA7185 sp900556545 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_B; Peptococcia; Peptococcales; Peptococcaceae; UBA7185; UBA7185 sp900556545 | |||||||||||
| CAZyme ID | MGYG000000285_00065 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11055; End: 12695 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 290 | 421 | 2.4e-30 | 0.37623762376237624 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4124 | ManB2 | 1.63e-13 | 275 | 467 | 140 | 328 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| pfam02156 | Glyco_hydro_26 | 3.64e-13 | 275 | 402 | 123 | 242 | Glycosyl hydrolase family 26. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QXM05316.1 | 1.80e-142 | 2 | 495 | 3 | 499 |
| QOX62356.1 | 2.25e-141 | 37 | 495 | 54 | 507 |
| QZY55773.1 | 5.62e-140 | 30 | 495 | 58 | 517 |
| QUH19438.1 | 9.03e-140 | 37 | 495 | 65 | 520 |
| AFS79156.1 | 4.62e-137 | 35 | 495 | 66 | 521 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VI0_A | 7.33e-33 | 216 | 479 | 12 | 275 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2BVD_A | 7.50e-33 | 216 | 479 | 12 | 272 | ChainA, ENDOGLUCANASE H [Acetivibrio thermocellus] |
| 2V3G_A | 7.50e-33 | 216 | 479 | 12 | 272 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2BV9_A | 8.81e-33 | 216 | 479 | 12 | 272 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2CIP_A | 1.91e-32 | 216 | 479 | 12 | 272 | ChainA, ENDOGLUCANASE H [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16218 | 7.83e-30 | 216 | 479 | 34 | 294 | Endoglucanase H OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celH PE=1 SV=1 |
| O05512 | 2.41e-07 | 312 | 472 | 185 | 329 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis (strain 168) OX=224308 GN=gmuG PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
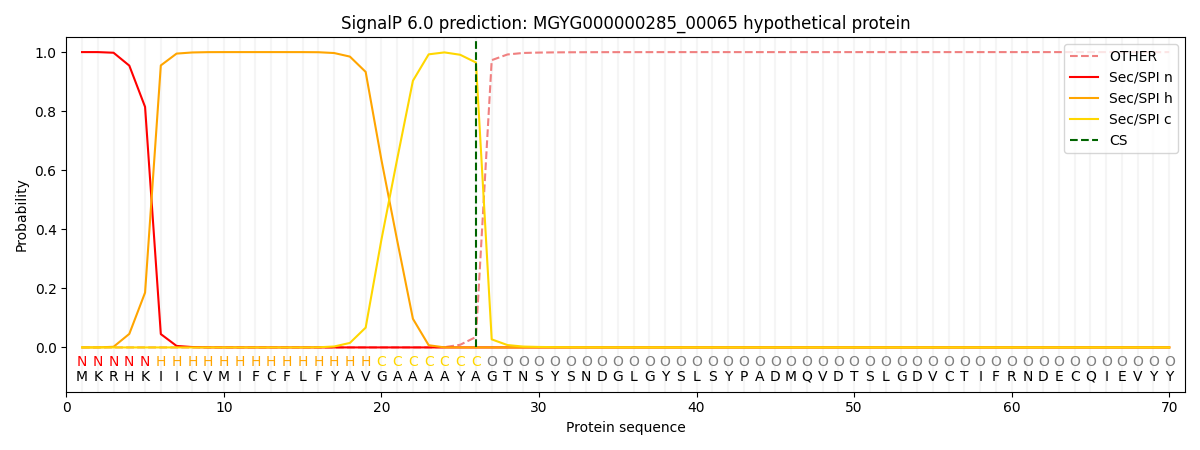
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000657 | 0.998339 | 0.000256 | 0.000274 | 0.000244 | 0.000219 |