You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000287_00539
You are here: Home > Sequence: MGYG000000287_00539
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
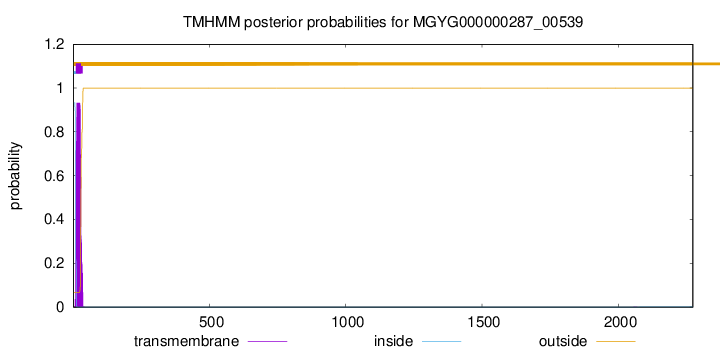
TMHMM annotations
Basic Information help
| Species | Robinsoniella sp900539655 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Robinsoniella; Robinsoniella sp900539655 | |||||||||||
| CAZyme ID | MGYG000000287_00539 | |||||||||||
| CAZy Family | GH141 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 59; End: 6883 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH141 | 44 | 592 | 1.2e-54 | 0.9886148007590133 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5492 | YjdB | 1.45e-20 | 1714 | 1906 | 144 | 324 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG5492 | YjdB | 3.34e-17 | 1481 | 1644 | 178 | 329 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG5492 | YjdB | 2.47e-16 | 1832 | 1994 | 177 | 326 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| COG5492 | YjdB | 6.73e-16 | 1516 | 1728 | 130 | 324 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
| pfam02368 | Big_2 | 5.00e-15 | 1486 | 1561 | 2 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWW32784.1 | 6.73e-112 | 42 | 802 | 30 | 755 |
| QWG02190.1 | 1.17e-51 | 1481 | 2098 | 322 | 904 |
| QWG06295.1 | 9.67e-50 | 1481 | 2002 | 322 | 892 |
| ANQ50384.1 | 2.97e-49 | 1481 | 2091 | 322 | 898 |
| ART28000.1 | 5.15e-49 | 1481 | 2091 | 322 | 897 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P33747 | 1.70e-16 | 1482 | 1639 | 35 | 190 | Uncharacterized protein CA_P0160 OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=CA_P0160 PE=3 SV=2 |
| P85991 | 8.78e-12 | 1829 | 2002 | 88 | 257 | Ig-like virion protein OS=Serratia phage KSP90 OX=552528 PE=1 SV=2 |
| A0A3R0A696 | 2.63e-07 | 1586 | 1700 | 288 | 399 | Alpha-L-arabinofuranosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=blArafA PE=1 SV=1 |
| P35838 | 3.06e-07 | 1837 | 1916 | 237 | 316 | Uncharacterized protein CA_C0552 OS=Clostridium acetobutylicum (strain ATCC 824 / DSM 792 / JCM 1419 / LMG 5710 / VKM B-1787) OX=272562 GN=CA_C0552 PE=4 SV=2 |
| Q086E4 | 9.61e-07 | 1449 | 1903 | 97 | 547 | Ice-binding protein 1 OS=Shewanella frigidimarina (strain NCIMB 400) OX=318167 GN=Sfri_1018 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
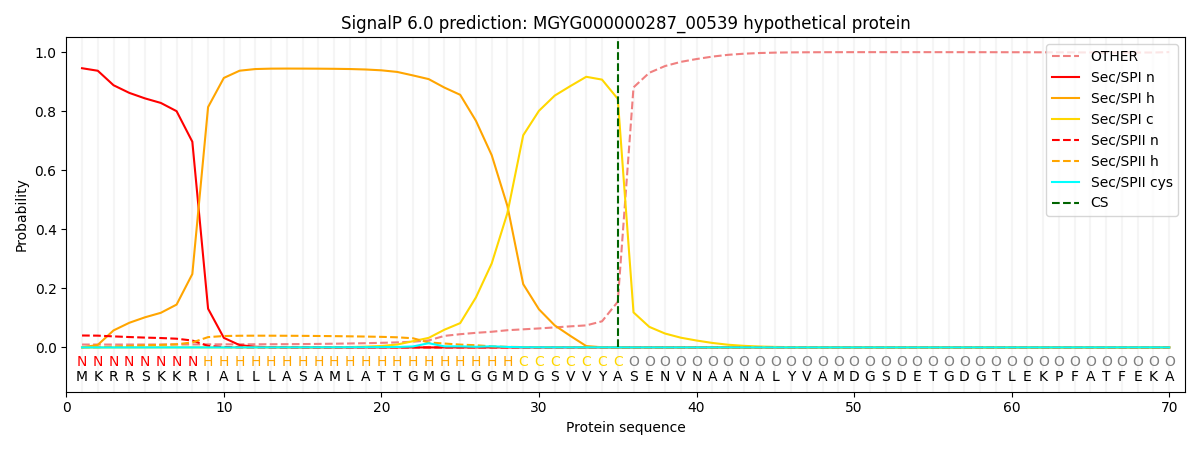
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.012388 | 0.939809 | 0.041628 | 0.005383 | 0.000466 | 0.000286 |