You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000287_00926
You are here: Home > Sequence: MGYG000000287_00926
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
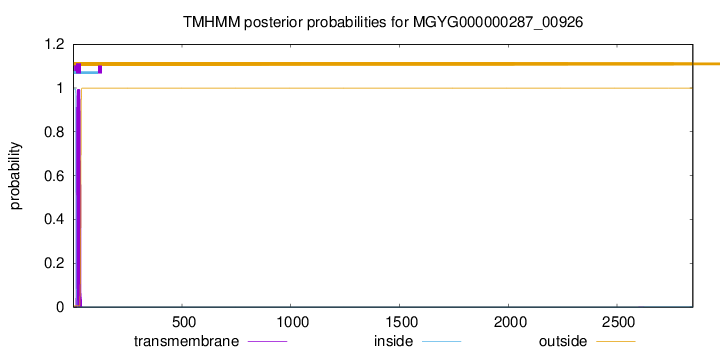
TMHMM annotations
Basic Information help
| Species | Robinsoniella sp900539655 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Robinsoniella; Robinsoniella sp900539655 | |||||||||||
| CAZyme ID | MGYG000000287_00926 | |||||||||||
| CAZy Family | CBM40 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 67057; End: 75606 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 1323 | 1760 | 3.3e-89 | 0.9532163742690059 |
| CBM40 | 1141 | 1297 | 1.5e-35 | 0.8994413407821229 |
| CBM51 | 2105 | 2240 | 5.9e-20 | 0.8955223880597015 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 5.80e-78 | 1320 | 1765 | 1 | 339 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| COG4409 | NanH | 3.76e-33 | 1142 | 1753 | 87 | 700 | Neuraminidase (sialidase) [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| pfam08305 | NPCBM | 1.13e-22 | 2089 | 2240 | 1 | 135 | NPCBM/NEW2 domain. This novel putative carbohydrate binding module (NPCBM) domain is found at the N-terminus of glycosyl hydrolase family 98 proteins. This domain has also been called the NEW2 domain (Naumoff DG. Phylogenetic analysis of alpha-galactosidases of the GH27 family. Molecular Biology (Engl Transl). (2004)38:388-399.) |
| cd00229 | SGNH_hydrolase | 1.02e-20 | 692 | 895 | 1 | 186 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| pfam13472 | Lipase_GDSL_2 | 3.34e-20 | 694 | 886 | 1 | 174 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ATD57534.1 | 1.77e-128 | 1143 | 1839 | 210 | 903 |
| QBJ75069.1 | 1.77e-128 | 1143 | 1839 | 210 | 903 |
| ATD54786.1 | 1.77e-128 | 1143 | 1839 | 210 | 903 |
| SLK16343.1 | 1.77e-128 | 1143 | 1839 | 210 | 903 |
| QJA08933.1 | 4.90e-123 | 1083 | 1836 | 13 | 752 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2BF6_A | 2.90e-97 | 1314 | 1771 | 4 | 448 | AtomicResolution Structure of the bacterial sialidase NanI from Clostridium perfringens in complex with alpha-Sialic Acid (Neu5Ac). [Clostridium perfringens] |
| 2VK5_A | 3.19e-97 | 1314 | 1771 | 4 | 448 | TheStructure Of Clostridium Perfringens Nani Sialidase And Its Catalytic Intermediates [Clostridium perfringens],2VK6_A The Structure Of Clostridium Perfringens Nani Sialidase And Its Catalytic Intermediates [Clostridium perfringens],2VK7_A The Structure Of Clostridium Perfringens Nani Sialidase And Its Catalytic Intermediates [Clostridium perfringens],2VK7_B The Structure Of Clostridium Perfringens Nani Sialidase And Its Catalytic Intermediates [Clostridium perfringens] |
| 5TSP_A | 5.72e-97 | 1310 | 1771 | 1 | 449 | Crystalstructure of the catalytic domain of Clostridium perfringens neuraminidase (NanI) in complex with a CHES [Clostridium perfringens ATCC 13124],5TSP_B Crystal structure of the catalytic domain of Clostridium perfringens neuraminidase (NanI) in complex with a CHES [Clostridium perfringens ATCC 13124] |
| 2VVZ_A | 1.25e-85 | 1333 | 1796 | 28 | 498 | Structureof the catalytic domain of Streptococcus pneumoniae sialidase NanA [Streptococcus pneumoniae],2VVZ_B Structure of the catalytic domain of Streptococcus pneumoniae sialidase NanA [Streptococcus pneumoniae] |
| 3H72_A | 1.23e-84 | 1310 | 1773 | 1 | 477 | Crystalstructure of Streptococcus pneumoniae D39 neuraminidase A precursor (NanA) in complex with NANA [Streptococcus pneumoniae R6],3H72_B Crystal structure of Streptococcus pneumoniae D39 neuraminidase A precursor (NanA) in complex with NANA [Streptococcus pneumoniae R6],3H73_A Crystal structure of Streptococcus pneumoniae D39 neuraminidase A precursor (NanA) in complex with DANA [Streptococcus pneumoniae R6],3H73_B Crystal structure of Streptococcus pneumoniae D39 neuraminidase A precursor (NanA) in complex with DANA [Streptococcus pneumoniae R6] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P29767 | 2.78e-120 | 1120 | 1847 | 188 | 910 | Sialidase OS=Clostridium septicum OX=1504 PE=3 SV=1 |
| P62575 | 1.84e-90 | 1117 | 1836 | 116 | 849 | Sialidase A OS=Streptococcus pneumoniae OX=1313 GN=nanA PE=1 SV=1 |
| P62576 | 1.84e-90 | 1117 | 1836 | 116 | 849 | Sialidase A OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=nanA PE=1 SV=1 |
| Q27701 | 4.60e-67 | 1123 | 1753 | 85 | 729 | Anhydrosialidase OS=Macrobdella decora OX=6405 PE=1 SV=1 |
| Q54727 | 2.90e-36 | 1119 | 1753 | 37 | 669 | Sialidase B OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=nanB PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
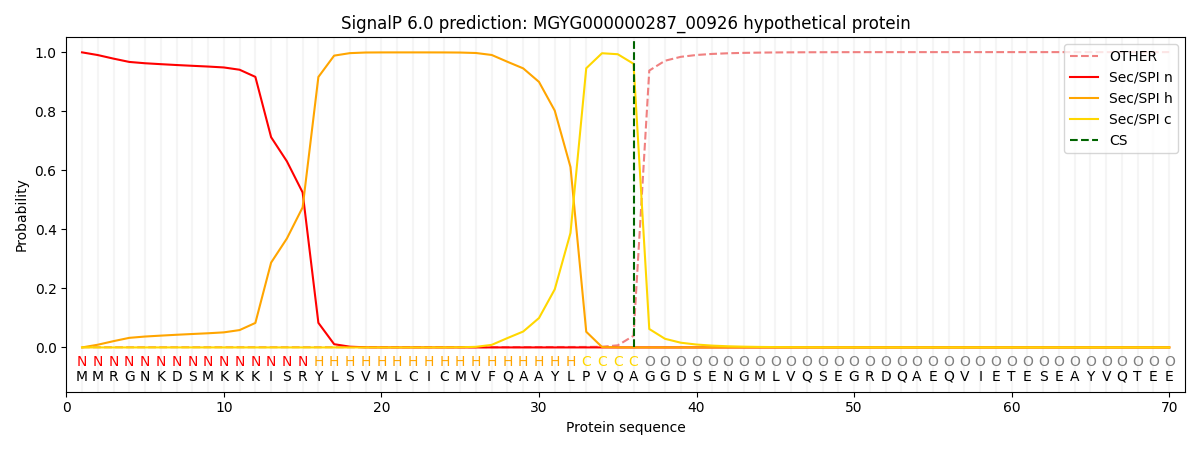
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001431 | 0.997395 | 0.000583 | 0.000183 | 0.000182 | 0.000173 |