You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000287_01183
You are here: Home > Sequence: MGYG000000287_01183
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Robinsoniella sp900539655 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Robinsoniella; Robinsoniella sp900539655 | |||||||||||
| CAZyme ID | MGYG000000287_01183 | |||||||||||
| CAZy Family | GH129 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 45721; End: 48066 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH129 | 437 | 768 | 1.6e-33 | 0.46116504854368934 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam11308 | Glyco_hydro_129 | 9.44e-131 | 413 | 738 | 1 | 324 | Glycosyl hydrolases related to GH101 family, GH129. This family of bacterial and lower eukaryote glycosyl hydrolases is related to CAZy family GH129,and distantly to GH101, and is made up of sub-families GHL1-GHL3. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QSW20179.1 | 3.29e-257 | 15 | 779 | 5 | 764 |
| AOH57445.1 | 5.35e-257 | 4 | 780 | 2 | 767 |
| QFF99969.1 | 2.46e-255 | 19 | 779 | 17 | 766 |
| CDH90420.1 | 1.40e-254 | 4 | 780 | 2 | 766 |
| QIC45839.1 | 1.40e-254 | 4 | 780 | 2 | 767 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OPQ_A | 8.71e-07 | 39 | 278 | 81 | 343 | A3,6-anhydro-D-galactosidase produced by Zobellia galactanivorans. This is an exo-lytic enzyme that hydrolyzes terminal 3,6-anhydro-D-galactose from the non-reducing end of carrageenan oligosaccharides. [Zobellia galactanivorans],5OPQ_B A 3,6-anhydro-D-galactosidase produced by Zobellia galactanivorans. This is an exo-lytic enzyme that hydrolyzes terminal 3,6-anhydro-D-galactose from the non-reducing end of carrageenan oligosaccharides. [Zobellia galactanivorans],5OPQ_C A 3,6-anhydro-D-galactosidase produced by Zobellia galactanivorans. This is an exo-lytic enzyme that hydrolyzes terminal 3,6-anhydro-D-galactose from the non-reducing end of carrageenan oligosaccharides. [Zobellia galactanivorans],5OPQ_D A 3,6-anhydro-D-galactosidase produced by Zobellia galactanivorans. This is an exo-lytic enzyme that hydrolyzes terminal 3,6-anhydro-D-galactose from the non-reducing end of carrageenan oligosaccharides. [Zobellia galactanivorans] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
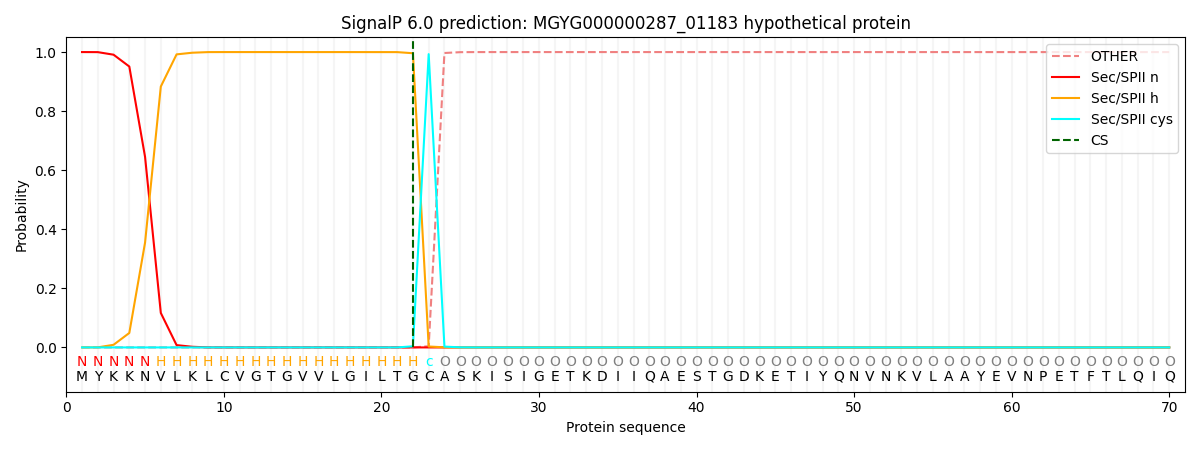
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000074 | 0.000000 | 0.000000 | 0.000000 |
