You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000287_01351
You are here: Home > Sequence: MGYG000000287_01351
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Robinsoniella sp900539655 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Robinsoniella; Robinsoniella sp900539655 | |||||||||||
| CAZyme ID | MGYG000000287_01351 | |||||||||||
| CAZy Family | GH123 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 52497; End: 58277 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH123 | 264 | 808 | 9e-163 | 0.9609665427509294 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13320 | DUF4091 | 5.98e-20 | 712 | 776 | 1 | 62 | Domain of unknown function (DUF4091). This presumed domain is functionally uncharacterized. This domain family is found in bacteria, archaea and eukaryotes, and is approximately 70 amino acids in length. There is a single completely conserved residue G that may be functionally important. |
| pfam13385 | Laminin_G_3 | 2.91e-19 | 993 | 1123 | 16 | 151 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| cd15482 | Sialidase_non-viral | 3.19e-10 | 1267 | 1550 | 47 | 332 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| NF033186 | internalin_K | 7.30e-09 | 1705 | 1755 | 510 | 560 | class 1 internalin InlK. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface-anchored proteins with an N-terminal signal peptide, leucine-rich repeats, and a C-terminal LPXTG processing and cell surface anchoring site. Members of this family are internalin K (InlK), a virulence factor. See articles PMID:17764999. for a general discussion of internalins, and PMID:21829365, PMID:22082958, and PMID:23958637 for more information about internalin K. |
| pfam02368 | Big_2 | 6.31e-08 | 1154 | 1204 | 27 | 77 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBA55846.1 | 0.0 | 84 | 1685 | 71 | 1665 |
| QTQ18982.1 | 0.0 | 84 | 1685 | 92 | 1686 |
| AXM91086.1 | 0.0 | 84 | 1685 | 71 | 1666 |
| ADO53743.1 | 0.0 | 84 | 1685 | 92 | 1687 |
| VEG17315.1 | 0.0 | 84 | 1685 | 92 | 1686 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5FQE_A | 1.95e-99 | 264 | 840 | 30 | 608 | Thedetails of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQE_B The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQF_A The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQF_B The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FR0_A The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens] |
| 5FQG_A | 2.26e-98 | 264 | 840 | 30 | 608 | Thedetails of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens],5FQH_A The details of glycolipid glycan hydrolysis by the structural analysis of a family 123 glycoside hydrolase from Clostridium perfringens [Clostridium perfringens] |
| 5L7V_A | 6.12e-77 | 290 | 782 | 51 | 519 | ChainA, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7V_B Chain B, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482] |
| 5L7R_A | 8.90e-77 | 290 | 782 | 66 | 534 | ChainA, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7R_B Chain B, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7U_A Chain A, Glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7U_B Chain B, Glycoside hydrolase [Phocaeicola vulgatus ATCC 8482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A0A3R0A696 | 9.61e-06 | 188 | 394 | 282 | 490 | Alpha-L-arabinofuranosidase OS=Bifidobacterium longum subsp. longum (strain ATCC 15707 / DSM 20219 / JCM 1217 / NCTC 11818 / E194b) OX=565042 GN=blArafA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
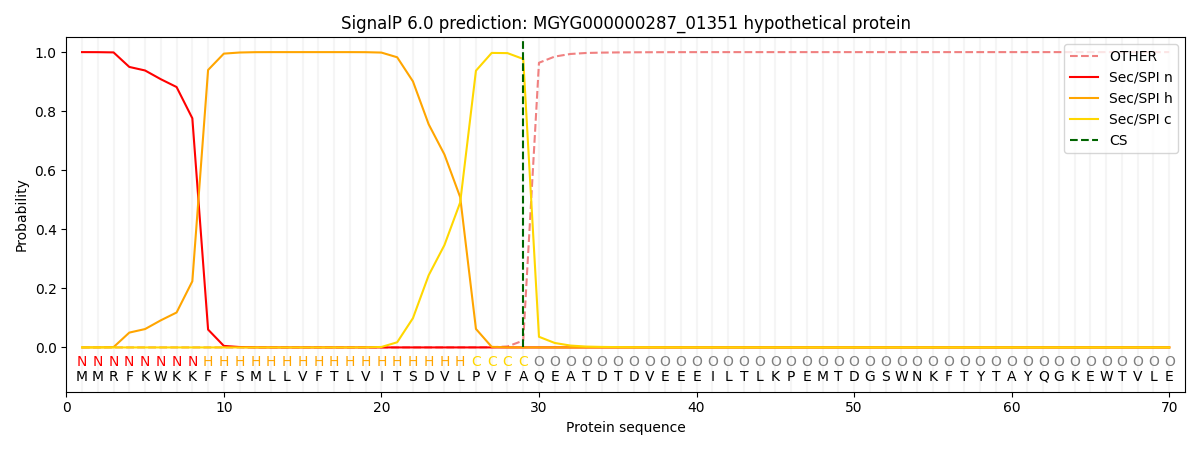
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000364 | 0.998857 | 0.000199 | 0.000203 | 0.000180 | 0.000158 |
