You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000287_03989
You are here: Home > Sequence: MGYG000000287_03989
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
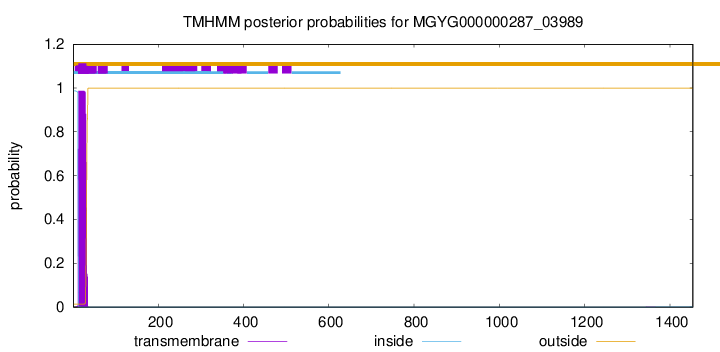
TMHMM annotations
Basic Information help
| Species | Robinsoniella sp900539655 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Robinsoniella; Robinsoniella sp900539655 | |||||||||||
| CAZyme ID | MGYG000000287_03989 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18708; End: 23072 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 115 | 310 | 1.4e-23 | 0.8425925925925926 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK15098 | PRK15098 | 3.30e-32 | 118 | 968 | 111 | 760 | beta-glucosidase BglX. |
| pfam01915 | Glyco_hydro_3_C | 2.69e-26 | 650 | 807 | 55 | 211 | Glycosyl hydrolase family 3 C-terminal domain. This domain is involved in catalysis and may be involved in binding beta-glucan. This domain is found associated with pfam00933. |
| pfam14310 | Fn3-like | 4.51e-20 | 890 | 962 | 1 | 71 | Fibronectin type III-like domain. This domain has a fibronectin type III-like structure. It is often found in association with pfam00933 and pfam01915. Its function is unknown. |
| PLN03080 | PLN03080 | 3.11e-18 | 688 | 959 | 488 | 769 | Probable beta-xylosidase; Provisional |
| cd04084 | CBM6_xylanase-like | 1.12e-13 | 1029 | 1150 | 20 | 123 | Carbohydrate Binding Module 6 (CBM6); many are appended to glycoside hydrolase (GH) family 11 and GH43 xylanase domains. This family includes carbohydrate binding module 6 (CBM6) domains that are appended mainly to glycoside hydrolase (GH) family domains, including GH3, GH11, and GH43 domains. These CBM6s are non-catalytic carbohydrate binding domains that facilitate the strong binding of the GH catalytic modules with their dedicated, insoluble substrates. Examples of proteins having CMB6s belonging to this family are Microbispora bispora GghA, a 1,4-beta-D-glucan glucohydrolase (GH3); Clostridium thermocellum xylanase U (GH11), and Penicillium purpurogenum ABF3, a bifunctional alpha-L-arabinofuranosidase/xylobiohydrolase (GH43). GH3 comprises enzymes with activities including beta-glucosidase (hydrolyzes beta-galactosidase) and beta-xylosidase (hydrolyzes 1,4-beta-D-xylosidase). GH11 family comprises enzymes with xylanase (endo-1,4-beta-xylanase) activity which catalyze the hydrolysis of beta-1,4 bonds of xylan, the major component of hemicelluloses, to generate xylooligosaccharides and xylose. GH43 includes beta-xylosidases and beta-xylanases, using aryl-glycosides as substrates. CBM6 is an unusual CBM as it represents a chimera of two distinct binding sites with different modes of binding: binding site I within the loop regions and binding site II on the concave face of the beta-sandwich fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QKS56187.1 | 7.56e-283 | 16 | 1150 | 16 | 1144 |
| AYA77556.1 | 3.81e-277 | 10 | 1150 | 12 | 1145 |
| QMV43973.1 | 2.13e-256 | 55 | 1156 | 21 | 1123 |
| AYQ75660.1 | 5.19e-231 | 19 | 1158 | 14 | 1140 |
| QOS79412.1 | 1.10e-199 | 38 | 1152 | 13 | 1125 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3AC0_A | 2.15e-27 | 111 | 966 | 47 | 829 | Crystalstructure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_B Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_C Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus],3AC0_D Crystal structure of Beta-glucosidase from Kluyveromyces marxianus in complex with glucose [Kluyveromyces marxianus] |
| 7MS2_A | 3.92e-27 | 661 | 973 | 379 | 668 | ChainA, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2],7MS2_B Chain B, Thermostable beta-glucosidase B [Acetivibrio thermocellus AD2] |
| 6JBS_A | 1.12e-24 | 703 | 965 | 527 | 795 | Bifunctionalxylosidase/glucosidase LXYL [Lentinula edodes],6JBS_B Bifunctional xylosidase/glucosidase LXYL [Lentinula edodes],6JBS_C Bifunctional xylosidase/glucosidase LXYL [Lentinula edodes],6JBS_D Bifunctional xylosidase/glucosidase LXYL [Lentinula edodes] |
| 4I3G_A | 1.16e-24 | 124 | 973 | 111 | 828 | CrystalStructure of DesR, a beta-glucosidase from Streptomyces venezuelae in complex with D-glucose. [Streptomyces venezuelae],4I3G_B Crystal Structure of DesR, a beta-glucosidase from Streptomyces venezuelae in complex with D-glucose. [Streptomyces venezuelae] |
| 3ABZ_A | 2.06e-24 | 111 | 966 | 47 | 829 | Crystalstructure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_B Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_C Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus],3ABZ_D Crystal structure of Se-Met labeled Beta-glucosidase from Kluyveromyces marxianus [Kluyveromyces marxianus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P27034 | 2.12e-27 | 100 | 965 | 39 | 802 | Beta-glucosidase OS=Rhizobium radiobacter OX=358 GN=cbg-1 PE=3 SV=1 |
| D5EY15 | 2.34e-27 | 122 | 962 | 80 | 849 | Xylan 1,4-beta-xylosidase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyl3A PE=1 SV=1 |
| P33363 | 1.27e-26 | 118 | 965 | 111 | 757 | Periplasmic beta-glucosidase OS=Escherichia coli (strain K12) OX=83333 GN=bglX PE=3 SV=2 |
| P14002 | 2.15e-26 | 661 | 973 | 379 | 668 | Thermostable beta-glucosidase B OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=bglB PE=1 SV=2 |
| A2QPK4 | 4.50e-25 | 703 | 973 | 486 | 750 | Probable beta-glucosidase D OS=Aspergillus niger (strain CBS 513.88 / FGSC A1513) OX=425011 GN=bglD PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
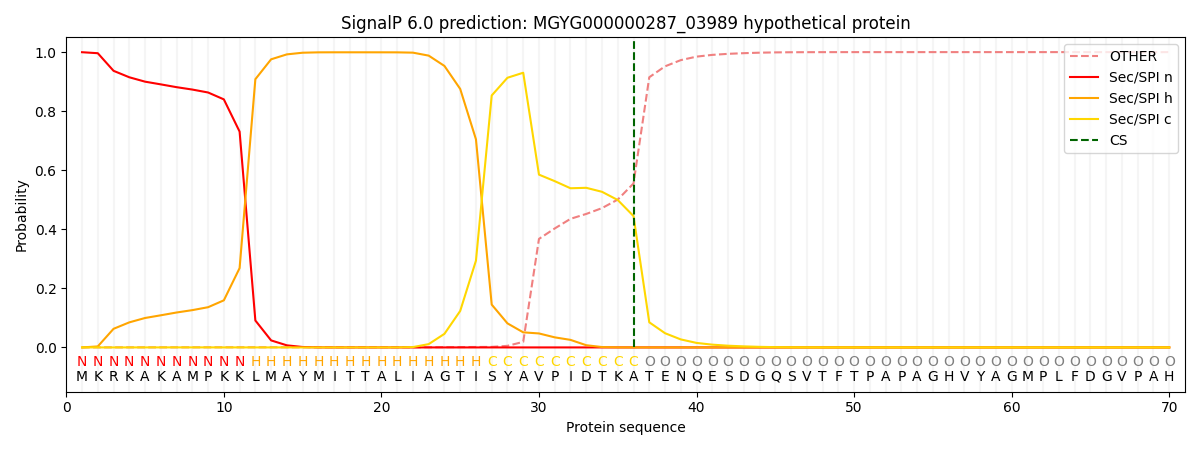
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001032 | 0.998015 | 0.000321 | 0.000225 | 0.000194 | 0.000173 |