You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000287_05502
You are here: Home > Sequence: MGYG000000287_05502
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
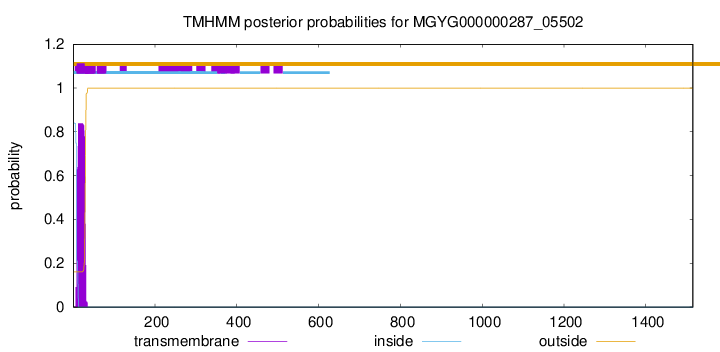
TMHMM annotations
Basic Information help
| Species | Robinsoniella sp900539655 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Robinsoniella; Robinsoniella sp900539655 | |||||||||||
| CAZyme ID | MGYG000000287_05502 | |||||||||||
| CAZy Family | GH51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5320; End: 9867 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH51 | 561 | 1228 | 5.3e-79 | 0.819047619047619 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3534 | AbfA | 1.28e-32 | 579 | 1228 | 40 | 498 | Alpha-L-arabinofuranosidase [Carbohydrate transport and metabolism]. |
| pfam06964 | Alpha-L-AF_C | 1.31e-18 | 850 | 939 | 1 | 93 | Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| smart00813 | Alpha-L-AF_C | 2.62e-15 | 850 | 1047 | 1 | 179 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| pfam13385 | Laminin_G_3 | 4.02e-12 | 91 | 263 | 2 | 151 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| COG5492 | YjdB | 3.81e-11 | 1200 | 1316 | 148 | 265 | Uncharacterized conserved protein YjdB, contains Ig-like domain [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AWB43005.1 | 1.34e-169 | 34 | 1226 | 3 | 1240 |
| AZH27584.1 | 5.02e-169 | 39 | 1226 | 8 | 1240 |
| ADL52276.1 | 2.07e-168 | 34 | 1226 | 9 | 1246 |
| BAV13155.1 | 2.07e-168 | 34 | 1226 | 9 | 1246 |
| QOH60128.1 | 1.90e-167 | 39 | 1226 | 8 | 1240 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ZPS_AAA | 7.99e-41 | 381 | 942 | 13 | 532 | ChainAAA, MgGH51 [Meripilus giganteus],6ZPV_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPW_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPX_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPY_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPZ_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ0_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ1_AAA Chain AAA, MgGH51 [Meripilus giganteus] |
| 2VRQ_A | 3.49e-15 | 588 | 774 | 51 | 203 | StructureOf An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus],2VRQ_B Structure Of An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus],2VRQ_C Structure Of An Inactive Mutant Of Arabinofuranosidase From Thermobacillus Xylanilyticus In Complex With A Pentasaccharide [Thermobacillus xylanilyticus] |
| 6ZT6_A | 6.11e-15 | 588 | 774 | 51 | 203 | ChainA, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT6_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT6_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_A Chain A, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT7_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus] |
| 6ZT8_A | 6.11e-15 | 588 | 774 | 51 | 203 | ChainA, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT8_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT8_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT9_A Chain A, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT9_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZT9_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus] |
| 6ZTA_A | 6.11e-15 | 588 | 774 | 51 | 203 | ChainA, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZTA_B Chain B, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus],6ZTA_C Chain C, Alpha-L-arabinofuranosidase [Thermobacillus xylanilyticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P82593 | 2.78e-121 | 366 | 1227 | 33 | 821 | Extracellular exo-alpha-L-arabinofuranosidase OS=Streptomyces chartreusis OX=1969 PE=1 SV=1 |
| Q9SG80 | 5.57e-51 | 378 | 939 | 52 | 553 | Alpha-L-arabinofuranosidase 1 OS=Arabidopsis thaliana OX=3702 GN=ASD1 PE=1 SV=1 |
| Q8VZR2 | 7.05e-51 | 381 | 939 | 54 | 552 | Alpha-L-arabinofuranosidase 2 OS=Arabidopsis thaliana OX=3702 GN=ASD2 PE=2 SV=1 |
| Q2U790 | 9.33e-44 | 376 | 937 | 33 | 536 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=abfA PE=3 SV=2 |
| B8NKA3 | 1.68e-43 | 376 | 937 | 33 | 536 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=abfA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
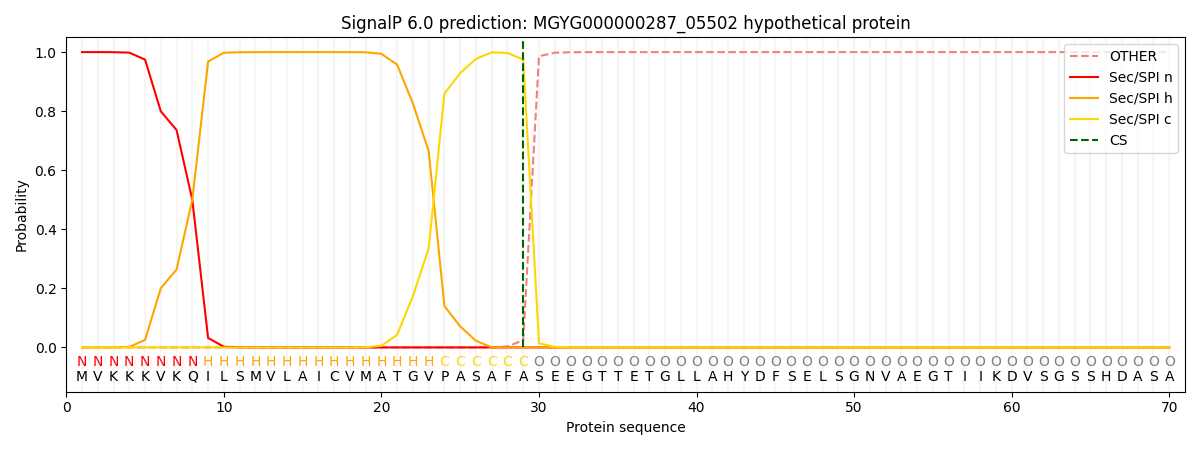
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000231 | 0.999069 | 0.000205 | 0.000179 | 0.000164 | 0.000146 |