You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000289_00816
You are here: Home > Sequence: MGYG000000289_00816
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella buccae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella buccae | |||||||||||
| CAZyme ID | MGYG000000289_00816 | |||||||||||
| CAZy Family | PL9 | |||||||||||
| CAZyme Description | Pectate lyase L | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 83259; End: 84704 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL9 | 21 | 418 | 2.2e-111 | 0.9813333333333333 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13229 | Beta_helix | 9.32e-10 | 158 | 320 | 6 | 157 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam13229 | Beta_helix | 0.001 | 144 | 259 | 39 | 138 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| pfam05048 | NosD | 0.004 | 118 | 327 | 7 | 178 | Periplasmic copper-binding protein (NosD). NosD is a periplasmic protein which is thought to insert copper into the exported reductase apoenzyme (NosZ). This region forms a parallel beta helix domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QMW86526.1 | 6.48e-158 | 3 | 435 | 1 | 437 |
| AAO79288.1 | 6.48e-158 | 3 | 435 | 1 | 437 |
| QQA07680.1 | 1.30e-157 | 3 | 435 | 1 | 437 |
| QUT42080.1 | 2.61e-157 | 3 | 435 | 1 | 437 |
| QUT69881.1 | 2.61e-157 | 3 | 435 | 1 | 437 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5OLQ_A | 2.51e-118 | 23 | 415 | 3 | 413 | Rhamnogalacturonanlyase [Bacteroides thetaiotaomicron],5OLQ_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLQ_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_B Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLR_C Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron],5OLS_A Rhamnogalacturonan lyase [Bacteroides thetaiotaomicron] |
| 1RU4_A | 1.11e-45 | 20 | 415 | 12 | 369 | ChainA, Pectate lyase [Dickeya chrysanthemi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0C1A6 | 5.34e-46 | 20 | 415 | 37 | 394 | Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
| P0C1A7 | 1.03e-44 | 20 | 415 | 37 | 394 | Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
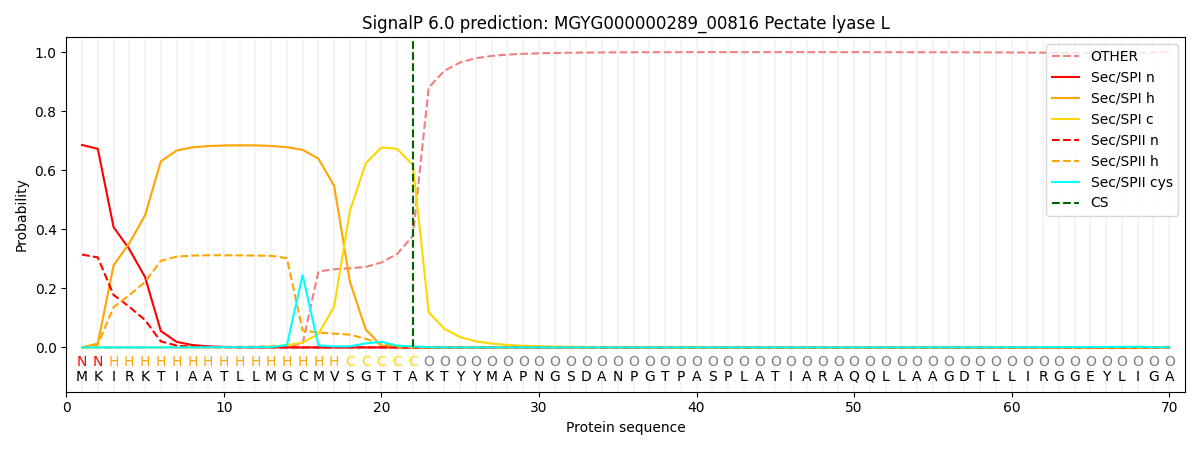
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000810 | 0.673254 | 0.325059 | 0.000320 | 0.000289 | 0.000245 |
