You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000289_01514
You are here: Home > Sequence: MGYG000000289_01514
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella buccae | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella buccae | |||||||||||
| CAZyme ID | MGYG000000289_01514 | |||||||||||
| CAZy Family | CE6 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 37667; End: 39895 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE6 | 346 | 458 | 1.1e-28 | 0.9797979797979798 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03629 | SASA | 3.02e-36 | 282 | 531 | 1 | 225 | Carbohydrate esterase, sialic acid-specific acetylesterase. The catalytic triad of this esterase enzyme comprises residues Ser127, His403 and Asp391 in UniProtKB:P70665. |
| COG3509 | LpqC | 5.00e-35 | 24 | 275 | 38 | 311 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| pfam10503 | Esterase_phd | 7.47e-13 | 38 | 166 | 6 | 146 | Esterase PHB depolymerase. This family of proteins include acetyl xylan esterases (AXE), feruloyl esterases (FAE), and poly(3-hydroxybutyrate) (PHB) depolymerases. |
| TIGR01840 | esterase_phb | 1.97e-11 | 38 | 182 | 4 | 156 | esterase, PHB depolymerase family. This model describes a subfamily among lipases of the ab-hydrolase family. This subfamily includes bacterial depolymerases for poly(3-hydroxybutyrate) (PHB) and related polyhydroxyalkanoates (PHA), as well as acetyl xylan esterases, feruloyl esterases, and others from fungi. [Fatty acid and phospholipid metabolism, Degradation] |
| COG4099 | COG4099 | 2.33e-04 | 38 | 175 | 179 | 328 | Predicted peptidase [General function prediction only]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNT67597.1 | 1.98e-118 | 279 | 538 | 31 | 288 |
| ADU86930.1 | 1.65e-104 | 273 | 742 | 4 | 579 |
| ALJ61522.1 | 4.06e-96 | 282 | 554 | 22 | 293 |
| QDO71565.1 | 8.88e-96 | 282 | 554 | 9 | 280 |
| QUT92908.1 | 2.07e-95 | 282 | 554 | 22 | 293 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MGQ_A | 2.01e-09 | 555 | 686 | 29 | 147 | PbXyn10CCBM APO [Prevotella bryantii B14] |
| 5X6S_A | 4.00e-08 | 38 | 154 | 22 | 148 | Acetylxylan esterase from Aspergillus awamori [Aspergillus awamori],5X6S_B Acetyl xylan esterase from Aspergillus awamori [Aspergillus awamori] |
| 3WYD_A | 1.93e-07 | 38 | 173 | 26 | 165 | C-terminalesterase domain of LC-Est1 [uncultured organism],3WYD_B C-terminal esterase domain of LC-Est1 [uncultured organism] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EXZ4 | 7.01e-65 | 279 | 538 | 42 | 299 | Carbohydrate acetyl esterase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=axe1-6A PE=1 SV=1 |
| B8YG19 | 6.98e-39 | 38 | 282 | 63 | 291 | Bifunctional acetylxylan esterase/xylanase XynS20E OS=Neocallimastix patriciarum OX=4758 GN=xynS20E PE=1 SV=1 |
| Q9Y871 | 2.28e-18 | 44 | 273 | 305 | 535 | Feruloyl esterase B OS=Piromyces equi OX=99929 GN=ESTA PE=2 SV=1 |
| A1CC33 | 6.56e-13 | 39 | 183 | 51 | 202 | Probable feruloyl esterase C OS=Aspergillus clavatus (strain ATCC 1007 / CBS 513.65 / DSM 816 / NCTC 3887 / NRRL 1 / QM 1276 / 107) OX=344612 GN=faeC-2 PE=3 SV=1 |
| A4D9B6 | 1.73e-11 | 33 | 268 | 45 | 269 | Probable feruloyl esterase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=faeC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
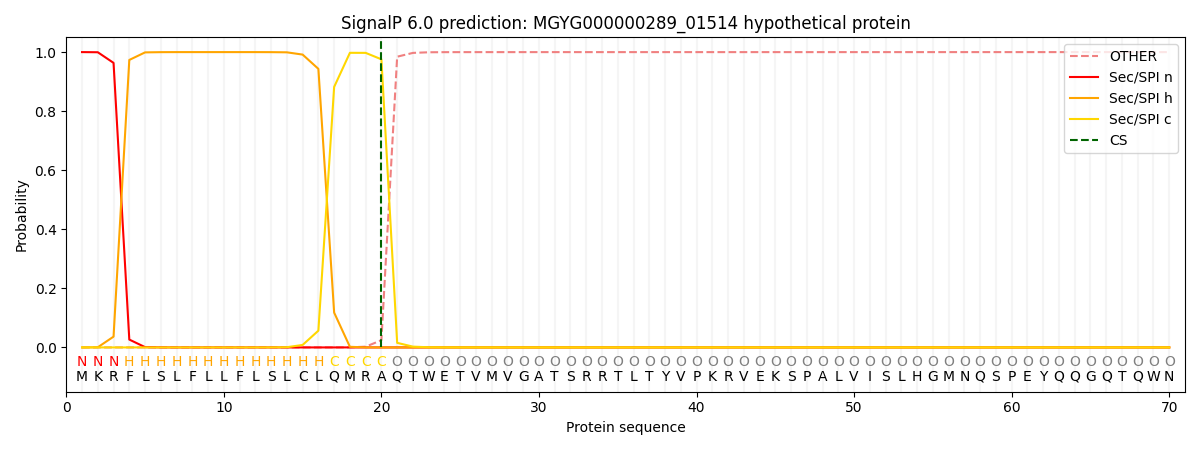
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000215 | 0.999115 | 0.000227 | 0.000147 | 0.000139 | 0.000136 |
