You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000319_01568
You are here: Home > Sequence: MGYG000000319_01568
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Pygmaiobacter massiliensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Pygmaiobacter; Pygmaiobacter massiliensis | |||||||||||
| CAZyme ID | MGYG000000319_01568 | |||||||||||
| CAZy Family | GT39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2030; End: 5074 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT39 | 603 | 822 | 1.1e-55 | 0.9282511210762332 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16192 | PMT_4TMC | 4.81e-24 | 832 | 1007 | 1 | 198 | C-terminal four TMM region of protein-O-mannosyltransferase. PMT_4TMC is the C-terminal four membrane-pass region of protein-O-mannosyltransferases and similar enzymes. |
| COG1928 | PMT1 | 1.86e-17 | 632 | 833 | 90 | 278 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| COG1928 | PMT1 | 1.73e-16 | 831 | 1012 | 498 | 696 | Dolichyl-phosphate-mannose--protein O-mannosyl transferase [Posttranslational modification, protein turnover, chaperones]. |
| COG5542 | COG5542 | 9.71e-13 | 64 | 345 | 92 | 353 | Mannosyltransferase related to Gpi18 [Carbohydrate transport and metabolism]. |
| COG4346 | COG4346 | 2.14e-12 | 625 | 871 | 129 | 330 | Predicted membrane-bound dolichyl-phosphate-mannose-protein mannosyltransferase [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWT55655.1 | 2.27e-191 | 22 | 1014 | 245 | 1256 |
| AIQ44967.1 | 3.14e-191 | 12 | 1011 | 5 | 1025 |
| AIQ50579.1 | 6.28e-187 | 12 | 1011 | 5 | 1025 |
| AIQ15817.1 | 2.07e-186 | 21 | 1011 | 18 | 1031 |
| AIQ66842.1 | 4.85e-184 | 21 | 1011 | 18 | 1031 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| L8F4Z2 | 1.60e-35 | 601 | 1009 | 59 | 515 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycolicibacterium smegmatis (strain MKD8) OX=1214915 GN=pmt PE=3 SV=1 |
| P9WN04 | 5.01e-31 | 609 | 1009 | 73 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain CDC 1551 / Oshkosh) OX=83331 GN=pmt PE=3 SV=2 |
| P9WN05 | 5.01e-31 | 609 | 1009 | 73 | 521 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Mycobacterium tuberculosis (strain ATCC 25618 / H37Rv) OX=83332 GN=pmt PE=1 SV=2 |
| Q8NRZ6 | 5.42e-23 | 632 | 1009 | 99 | 519 | Probable dolichyl-phosphate-mannose--protein mannosyltransferase OS=Corynebacterium glutamicum (strain ATCC 13032 / DSM 20300 / BCRC 11384 / JCM 1318 / LMG 3730 / NCIMB 10025) OX=196627 GN=pmt PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
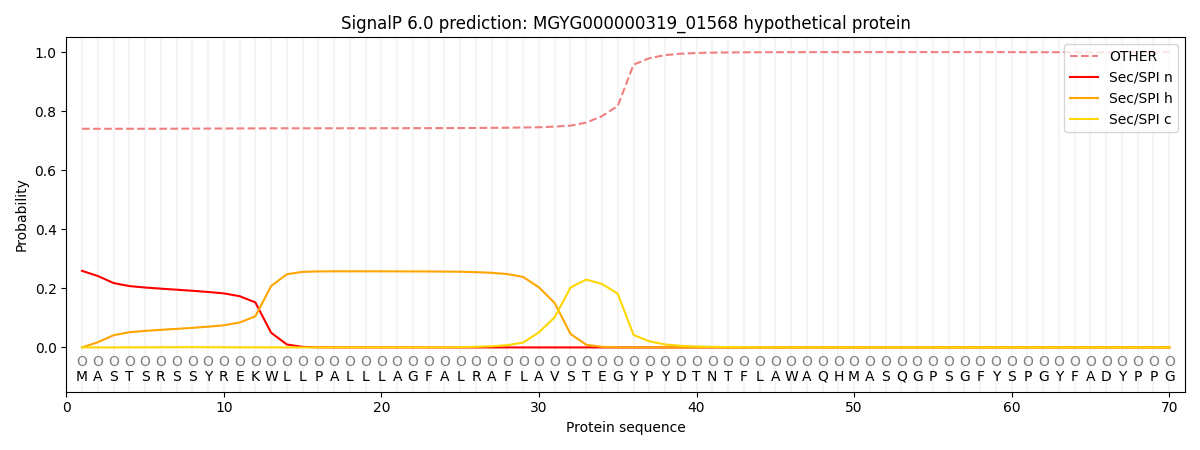
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.748270 | 0.249700 | 0.001039 | 0.000318 | 0.000253 | 0.000430 |
TMHMM Annotations download full data without filtering help
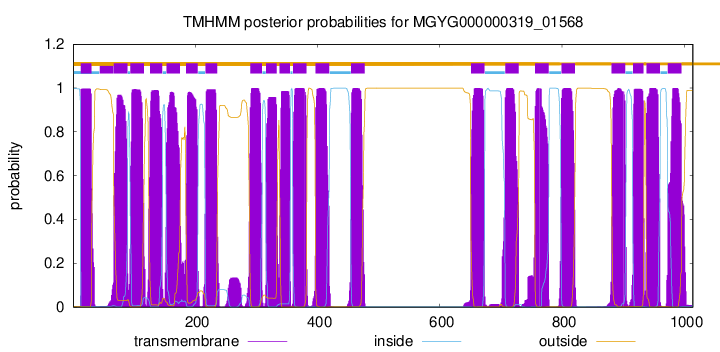
| start | end |
|---|---|
| 13 | 30 |
| 67 | 89 |
| 94 | 116 |
| 126 | 146 |
| 153 | 175 |
| 185 | 204 |
| 217 | 236 |
| 290 | 309 |
| 316 | 333 |
| 338 | 355 |
| 360 | 382 |
| 397 | 419 |
| 455 | 477 |
| 651 | 673 |
| 707 | 729 |
| 756 | 778 |
| 799 | 821 |
| 881 | 903 |
| 916 | 933 |
| 938 | 960 |
| 973 | 995 |
