You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000338_01642
You are here: Home > Sequence: MGYG000000338_01642
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
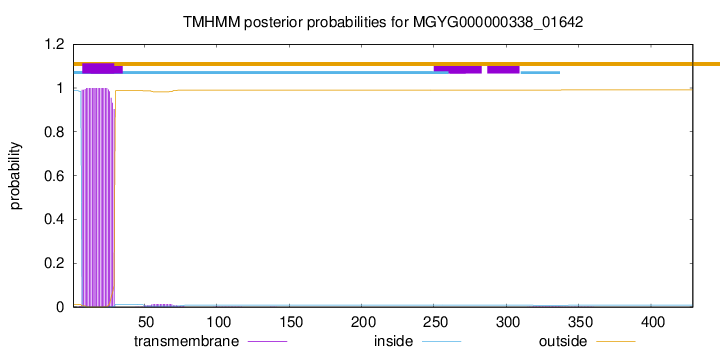
TMHMM annotations
Basic Information help
| Species | Anaerobiospirillum sp900543125 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Succinivibrionaceae; Anaerobiospirillum; Anaerobiospirillum sp900543125 | |||||||||||
| CAZyme ID | MGYG000000338_01642 | |||||||||||
| CAZy Family | GT30 | |||||||||||
| CAZyme Description | 3-deoxy-D-manno-octulosonic acid transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1877; End: 3166 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT30 | 53 | 215 | 1.3e-53 | 0.9209039548022598 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK05749 | PRK05749 | 4.00e-126 | 7 | 423 | 1 | 419 | 3-deoxy-D-manno-octulosonic-acid transferase; Reviewed |
| COG1519 | KdtA | 1.43e-109 | 8 | 423 | 1 | 417 | 3-deoxy-D-manno-octulosonic-acid transferase [Cell wall/membrane/envelope biogenesis]. |
| pfam04413 | Glycos_transf_N | 3.33e-53 | 43 | 215 | 2 | 176 | 3-Deoxy-D-manno-octulosonic-acid transferase (kdotransferase). Members of this family transfer activated sugars to a variety of substrates, including glycogen, fructose-6-phosphate and lipopolysaccharides. Members of the family transfer UDP, ADP, GDP or CMP linked sugars. The Glycos_transf_N region is flanked at the N-terminus by a signal peptide and at the C-terminus by Glycos_transf_1 (pfam00534). The eukaryotic glycogen synthases may be distant members of this bacterial family. |
| pfam00534 | Glycos_transf_1 | 1.64e-05 | 236 | 403 | 6 | 155 | Glycosyl transferases group 1. Mutations in this domain of PIGA lead to disease (Paroxysmal Nocturnal haemoglobinuria). Members of this family transfer activated sugars to a variety of substrates, including glycogen, Fructose-6-phosphate and lipopolysaccharides. Members of this family transfer UDP, ADP, GDP or CMP linked sugars. The eukaryotic glycogen synthases may be distant members of this family. |
| cd03807 | GT4_WbnK-like | 0.006 | 138 | 270 | 80 | 227 | Shigella dysenteriae WbnK and similar proteins. This family is most closely related to the GT4 family of glycosyltransferases. WbnK in Shigella dysenteriae has been shown to be involved in the type 7 O-antigen biosynthesis. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QCF36950.1 | 5.34e-89 | 10 | 421 | 3 | 411 |
| QIR05043.1 | 1.19e-87 | 10 | 421 | 3 | 411 |
| ARR45740.1 | 4.09e-83 | 11 | 418 | 6 | 416 |
| CAE6880445.1 | 4.33e-83 | 11 | 418 | 6 | 413 |
| AHK19294.1 | 1.81e-82 | 11 | 425 | 5 | 420 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2XCI_A | 3.22e-41 | 53 | 390 | 37 | 344 | Membrane-embeddedmonofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form [Aquifex aeolicus],2XCI_B Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form [Aquifex aeolicus],2XCI_C Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form [Aquifex aeolicus],2XCI_D Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, substrate-free form [Aquifex aeolicus],2XCU_A Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP [Aquifex aeolicus],2XCU_B Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP [Aquifex aeolicus],2XCU_C Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP [Aquifex aeolicus],2XCU_D Membrane-embedded monofunctional glycosyltransferase WaaA of Aquifex aeolicus, complex with CMP [Aquifex aeolicus] |
| 4BFC_A | 2.20e-18 | 237 | 429 | 44 | 235 | ChainA, 3-DEOXY-D-MANNO-OCTULOSONIC-ACID TRANSFERASE [Acinetobacter baumannii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0AC77 | 1.74e-73 | 11 | 425 | 5 | 420 | 3-deoxy-D-manno-octulosonic acid transferase OS=Escherichia coli O157:H7 OX=83334 GN=waaA PE=3 SV=1 |
| P0AC76 | 1.74e-73 | 11 | 425 | 5 | 420 | 3-deoxy-D-manno-octulosonic acid transferase OS=Escherichia coli O6:H1 (strain CFT073 / ATCC 700928 / UPEC) OX=199310 GN=waaA PE=3 SV=1 |
| P0AC75 | 1.74e-73 | 11 | 425 | 5 | 420 | 3-deoxy-D-manno-octulosonic acid transferase OS=Escherichia coli (strain K12) OX=83333 GN=waaA PE=1 SV=1 |
| P44806 | 3.66e-69 | 12 | 428 | 6 | 426 | 3-deoxy-D-manno-octulosonic acid transferase OS=Haemophilus influenzae (strain ATCC 51907 / DSM 11121 / KW20 / Rd) OX=71421 GN=waaA PE=1 SV=1 |
| Q45374 | 1.35e-45 | 56 | 425 | 55 | 427 | 3-deoxy-D-manno-octulosonic acid transferase OS=Bordetella pertussis OX=520 GN=waaA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
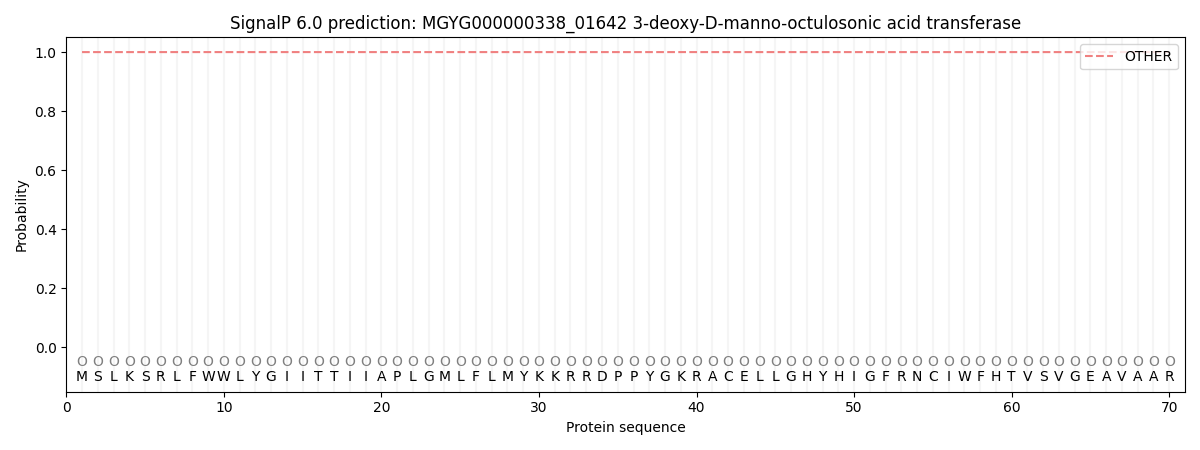
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000036 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |