You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000338_01651
Basic Information
help
| Species |
Anaerobiospirillum sp900543125
|
| Lineage |
Bacteria; Proteobacteria; Gammaproteobacteria; Enterobacterales; Succinivibrionaceae; Anaerobiospirillum; Anaerobiospirillum sp900543125
|
| CAZyme ID |
MGYG000000338_01651
|
| CAZy Family |
PL9 |
| CAZyme Description |
Pectate disaccharide-lyase
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 185 |
|
20350.32 |
6.0923 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000338 |
3410865 |
MAG |
Sweden |
Europe |
|
| Gene Location |
Start: 236;
End: 793
Strand: +
|
| Family |
Start |
End |
Evalue |
family coverage |
| PL9 |
1 |
182 |
9e-48 |
0.5013333333333333 |
MGYG000000338_01651 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 1RU4_A
|
1.77e-17 |
1 |
182 |
190 |
368 |
ChainA, Pectate lyase [Dickeya chrysanthemi] |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
P22751
|
1.10e-50 |
1 |
146 |
544 |
688 |
Pectate disaccharide-lyase OS=Dickeya chrysanthemi OX=556 GN=pelX PE=1 SV=1 |
|
P0C1A6
|
7.78e-17 |
1 |
182 |
215 |
393 |
Pectate lyase L OS=Dickeya chrysanthemi OX=556 GN=pelL PE=3 SV=1 |
|
P0C1A7
|
1.06e-16 |
1 |
182 |
215 |
393 |
Pectate lyase L OS=Dickeya dadantii (strain 3937) OX=198628 GN=pelL PE=1 SV=1 |
This protein is predicted as OTHER
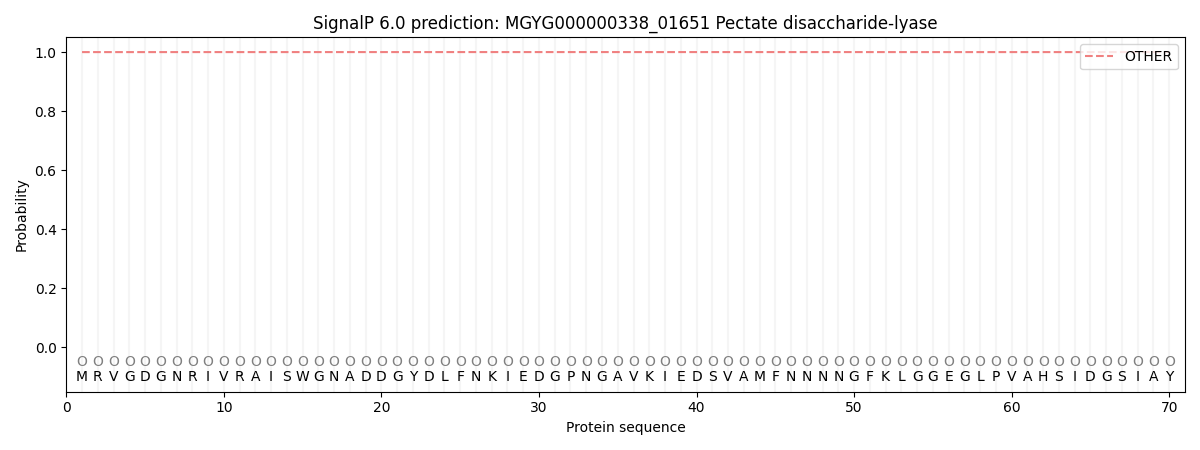
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
1.000059
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
There is no transmembrane helices in MGYG000000338_01651.

