You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000343_00221
You are here: Home > Sequence: MGYG000000343_00221
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Odoribacter sp900548135 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Marinifilaceae; Odoribacter; Odoribacter sp900548135 | |||||||||||
| CAZyme ID | MGYG000000343_00221 | |||||||||||
| CAZy Family | GH3 | |||||||||||
| CAZyme Description | Beta-hexosaminidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 293487; End: 297284 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH3 | 587 | 831 | 2e-72 | 0.9861111111111112 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| PRK10077 | xylE | 0.0 | 1 | 534 | 2 | 477 | D-xylose transporter XylE; Provisional |
| PLN03080 | PLN03080 | 2.34e-141 | 550 | 1227 | 37 | 755 | Probable beta-xylosidase; Provisional |
| cd17359 | MFS_XylE_like | 7.46e-140 | 12 | 516 | 2 | 383 | D-xylose-proton symporter and similar transporters of the Major Facilitator Superfamily. This subfamily includes bacterial transporters such as D-xylose-proton symporter (XylE or XylT), arabinose-proton symporter (AraE), galactose-proton symporter (GalP), major myo-inositol transporter IolT, glucose transport protein, putative metabolite transport proteins YfiG, YncC, and YwtG, and similar proteins. The symporters XylE, AraE, and GalP facilitate the uptake of D-xylose, arabinose, and galactose, respectively, across the boundary membrane with the concomitant transport of protons into the cell. IolT is involved in polyol metabolism and myo-inositol degradation into acetyl-CoA. The XylE-like subfamily belongs to the Glucose transporter -like (GLUT-like) family of the Major Facilitator Superfamily (MFS) of membrane transport proteins. MFS proteins are thought to function through a single substrate binding site, alternating-access mechanism involving a rocker-switch type of movement. |
| PRK15098 | PRK15098 | 2.59e-137 | 532 | 1252 | 1 | 762 | beta-glucosidase BglX. |
| TIGR00879 | SP | 1.52e-112 | 2 | 525 | 19 | 481 | MFS transporter, sugar porter (SP) family. This model represent the sugar porter subfamily of the major facilitator superfamily (pfam00083) [Transport and binding proteins, Carbohydrates, organic alcohols, and acids] |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QKJ63413.1 | 0.0 | 524 | 1265 | 2 | 741 |
| AOW10543.1 | 0.0 | 536 | 1265 | 22 | 748 |
| QQT29233.1 | 0.0 | 535 | 1265 | 14 | 744 |
| QQT54734.1 | 0.0 | 535 | 1265 | 14 | 744 |
| QRY59956.1 | 0.0 | 535 | 1265 | 14 | 744 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4JA3_A | 1.29e-161 | 2 | 530 | 3 | 479 | Partiallyoccluded inward open conformation of the xylose transporter XylE from E. coli [Escherichia coli K-12],4JA3_B Partially occluded inward open conformation of the xylose transporter XylE from E. coli [Escherichia coli K-12],4JA4_A Inward open conformation of the xylose transporter XylE from E. coli [Escherichia coli K-12],4JA4_B Inward open conformation of the xylose transporter XylE from E. coli [Escherichia coli K-12],4JA4_C Inward open conformation of the xylose transporter XylE from E. coli [Escherichia coli K-12] |
| 4GBY_A | 1.58e-161 | 2 | 530 | 3 | 479 | Thestructure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-xylose [Escherichia coli K-12],4GBZ_A The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to D-glucose [Escherichia coli K-12],4GC0_A The structure of the MFS (major facilitator superfamily) proton:xylose symporter XylE bound to 6-bromo-6-deoxy-D-glucose [Escherichia coli K-12] |
| 4QIQ_A | 2.91e-161 | 2 | 530 | 2 | 478 | Crystalstructure of D-xylose-proton symporter [Escherichia coli K-12] |
| 6N3I_A | 4.54e-159 | 2 | 530 | 30 | 506 | Crystalstructure of a double Trp XylE mutants (G58W/L315W) [Escherichia coli] |
| 7VC7_A | 2.39e-105 | 565 | 1201 | 25 | 677 | ChainA, xylan 1,4-beta-xylosidase [Phanerodontia chrysosporium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EY15 | 1.30e-177 | 534 | 1241 | 2 | 846 | Xylan 1,4-beta-xylosidase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyl3A PE=1 SV=1 |
| P0AGF5 | 8.66e-161 | 2 | 530 | 3 | 479 | D-xylose-proton symporter OS=Escherichia coli O157:H7 OX=83334 GN=xylE PE=3 SV=1 |
| P0AGF4 | 8.66e-161 | 2 | 530 | 3 | 479 | D-xylose-proton symporter OS=Escherichia coli (strain K12) OX=83333 GN=xylE PE=1 SV=1 |
| Q94KD8 | 3.39e-122 | 543 | 1241 | 32 | 750 | Probable beta-D-xylosidase 2 OS=Arabidopsis thaliana OX=3702 GN=BXL2 PE=2 SV=1 |
| B0Y0I4 | 7.09e-122 | 557 | 1260 | 44 | 757 | Probable exo-1,4-beta-xylosidase bxlB OS=Neosartorya fumigata (strain CEA10 / CBS 144.89 / FGSC A1163) OX=451804 GN=bxlB PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000014 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
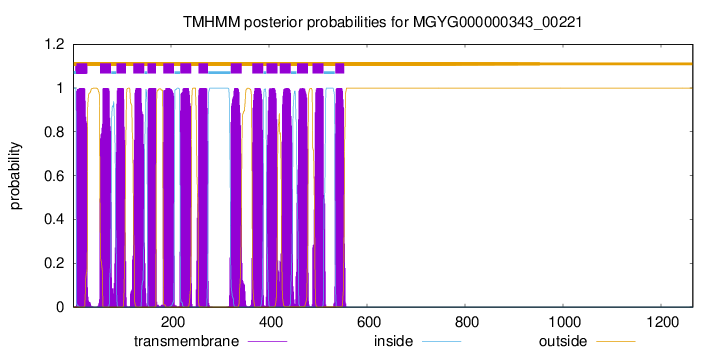
| start | end |
|---|---|
| 7 | 29 |
| 55 | 77 |
| 89 | 108 |
| 123 | 145 |
| 152 | 169 |
| 184 | 206 |
| 219 | 241 |
| 256 | 275 |
| 322 | 344 |
| 366 | 388 |
| 395 | 417 |
| 422 | 444 |
| 457 | 479 |
| 489 | 511 |
| 535 | 553 |
