You are browsing environment: HUMAN GUT
MGYG000000345_00960
Basic Information
help
Species
UBA1829 sp002338895
Lineage
Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA1829; UBA1829 sp002338895
CAZyme ID
MGYG000000345_00960
CAZy Family
GH42
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
1097
123343.91
9.6498
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000000345
3773289
MAG
Sweden
Europe
Gene Location
Start: 134635;
End: 137928
Strand: -
No EC number prediction in MGYG000000345_00960.
Family
Start
End
Evalue
family coverage
GH42
577
762
2.7e-17
0.48247978436657685
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam02449
Glyco_hydro_42
2.07e-09
578
769
122
312
Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues.
more
COG1874
GanA
3.52e-06
607
713
173
283
Beta-galactosidase GanA [Carbohydrate transport and metabolism].
more
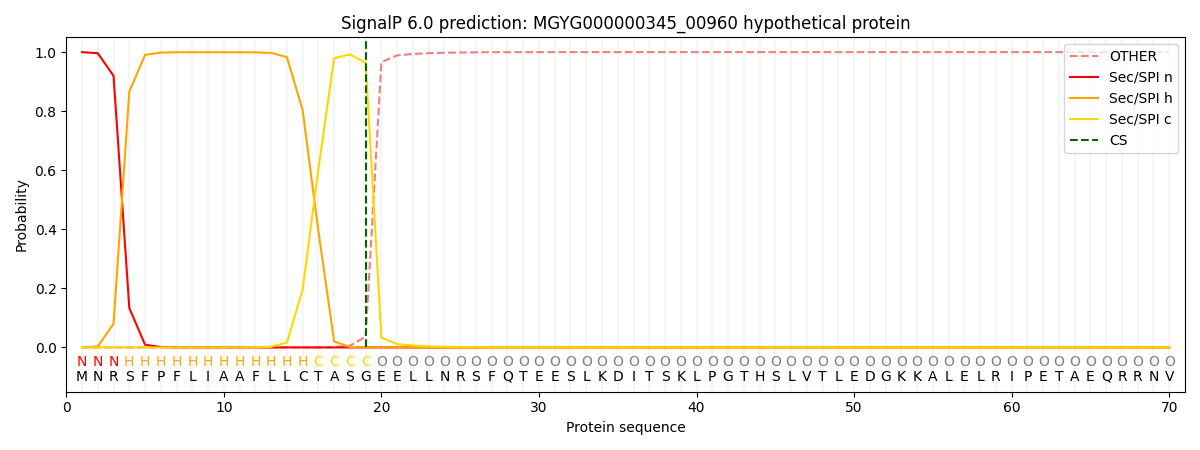
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000692
0.998203
0.000558
0.000181
0.000173
0.000165
There is no transmembrane helices in MGYG000000345_00960.