You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000345_01600
You are here: Home > Sequence: MGYG000000345_01600
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA1829 sp002338895 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA1829; UBA1829 sp002338895 | |||||||||||
| CAZyme ID | MGYG000000345_01600 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 64412; End: 66001 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 9.88e-19 | 218 | 489 | 24 | 271 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 6.01e-10 | 265 | 491 | 118 | 374 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44326.1 | 3.56e-162 | 18 | 517 | 30 | 517 |
| AVM44708.1 | 4.93e-106 | 14 | 518 | 16 | 499 |
| AVM45721.1 | 2.24e-79 | 33 | 519 | 27 | 516 |
| AVM43433.1 | 2.69e-79 | 51 | 518 | 47 | 510 |
| AVM46198.1 | 1.71e-77 | 41 | 518 | 19 | 501 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4LX4_A | 1.54e-09 | 213 | 401 | 33 | 214 | CrystalStructure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],4LX4_B Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],4LX4_C Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],4LX4_D Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],6R2J_A Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501],6R2J_B Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501],6R2J_C Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501],6R2J_D Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501] |
| 4EE9_A | 4.62e-09 | 213 | 468 | 24 | 273 | Crystalstructure of the RBcel1 endo-1,4-glucanase [uncultured bacterium],4M24_A Crystal structure of the endo-1,4-glucanase, RBcel1, in complex with cellobiose [uncultured bacterium] |
| 7P6G_A | 1.10e-08 | 213 | 468 | 24 | 273 | ChainA, Endoglucanase [uncultured bacterium],7P6G_B Chain B, Endoglucanase [uncultured bacterium],7P6H_A Chain A, Endoglucanase [uncultured bacterium],7P6H_B Chain B, Endoglucanase [uncultured bacterium] |
| 6ZZ3_A | 1.45e-08 | 213 | 468 | 24 | 273 | ChainA, Endoglucanase [uncultured bacterium],6ZZ3_B Chain B, Endoglucanase [uncultured bacterium],6ZZ3_C Chain C, Endoglucanase [uncultured bacterium],6ZZ3_D Chain D, Endoglucanase [uncultured bacterium] |
| 7P6I_A | 1.46e-08 | 213 | 468 | 24 | 273 | ChainA, Endoglucanase [uncultured bacterium],7P6J_A Chain A, Endoglucanase [uncultured bacterium],7P6J_B Chain B, Endoglucanase [uncultured bacterium],7P6J_C Chain C, Endoglucanase [uncultured bacterium],7P6J_D Chain D, Endoglucanase [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| W8QRE4 | 8.77e-06 | 268 | 452 | 116 | 296 | Beta-xylosidase OS=Phanerodontia chrysosporium OX=2822231 GN=Xyl5 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
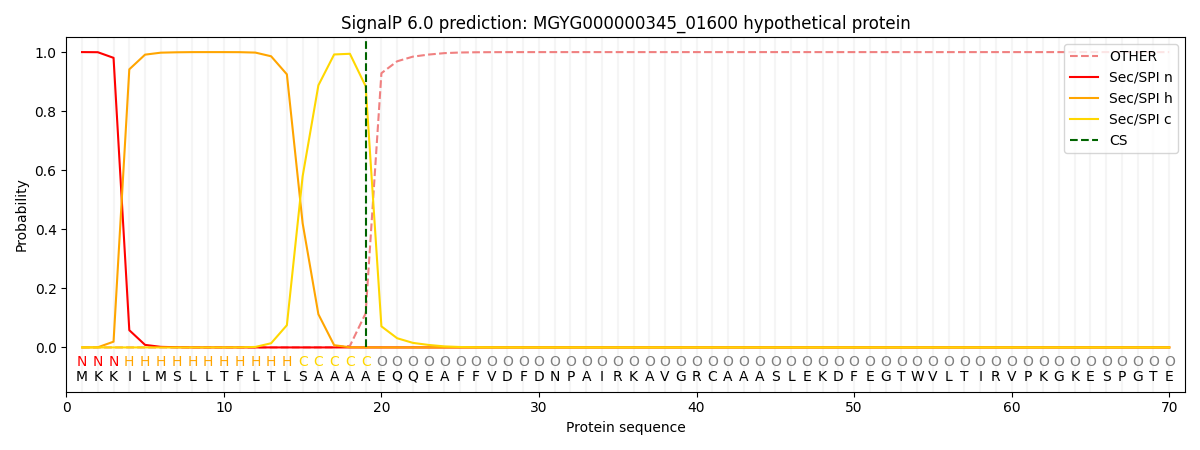
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000386 | 0.998922 | 0.000206 | 0.000178 | 0.000157 | 0.000153 |
