You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000345_02468
Basic Information
help
| Species |
UBA1829 sp002338895
|
| Lineage |
Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA1829; UBA1829 sp002338895
|
| CAZyme ID |
MGYG000000345_02468
|
| CAZy Family |
GH148 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 1085 |
|
122292 |
10.0117 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000345 |
3773289 |
MAG |
Sweden |
Europe |
|
| Gene Location |
Start: 5518;
End: 8775
Strand: -
|
No EC number prediction in MGYG000000345_02468.
| Family |
Start |
End |
Evalue |
family coverage |
| GH148 |
441 |
578 |
1.1e-23 |
0.9210526315789473 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG4817
|
GINS |
0.003 |
599 |
652 |
15 |
86 |
DNA-binding ferritin-like protein (Dps family) [Replication, recombination and repair]. |
This protein is predicted as SP
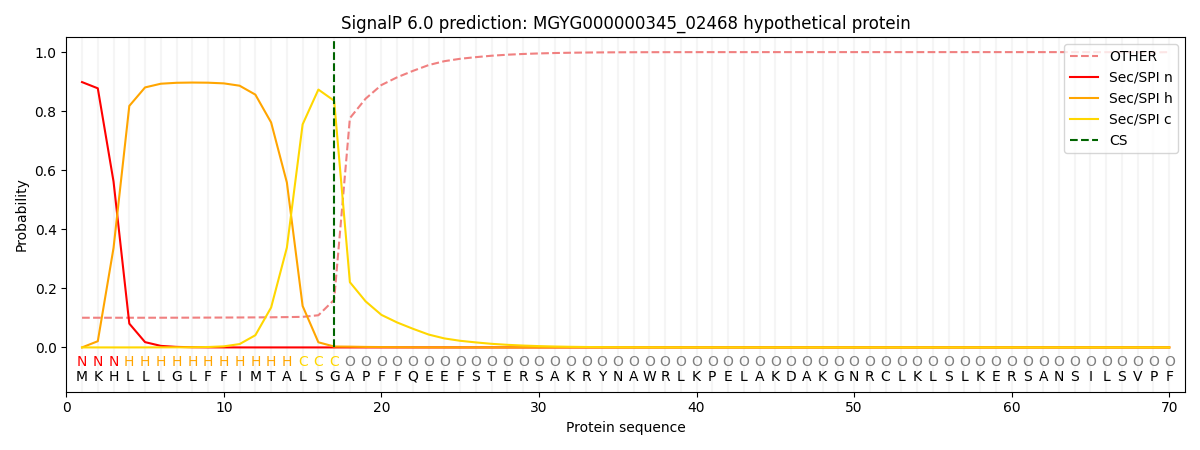
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.108305
|
0.889263
|
0.001519
|
0.000294
|
0.000291
|
0.000305
|
There is no transmembrane helices in MGYG000000345_02468.

