You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000345_02955
Basic Information
help
| Species |
UBA1829 sp002338895
|
| Lineage |
Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA1829; UBA1829 sp002338895
|
| CAZyme ID |
MGYG000000345_02955
|
| CAZy Family |
GH148 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 1098 |
|
124809.06 |
9.721 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000345 |
3773289 |
MAG |
Sweden |
Europe |
|
| Gene Location |
Start: 2366;
End: 5662
Strand: -
|
No EC number prediction in MGYG000000345_02955.
| Family |
Start |
End |
Evalue |
family coverage |
| GH148 |
484 |
635 |
2.9e-33 |
0.993421052631579 |
| GH148 |
692 |
809 |
3.6e-18 |
0.7631578947368421 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam00150
|
Cellulase |
0.002 |
479 |
564 |
3 |
85 |
Cellulase (glycosyl hydrolase family 5). |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
Q7Y223
|
7.80e-07 |
462 |
635 |
38 |
215 |
Mannan endo-1,4-beta-mannosidase 2 OS=Arabidopsis thaliana OX=3702 GN=MAN2 PE=2 SV=1 |
|
Q10B67
|
1.91e-06 |
481 |
639 |
36 |
200 |
Mannan endo-1,4-beta-mannosidase 4 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN4 PE=2 SV=2 |
This protein is predicted as SP
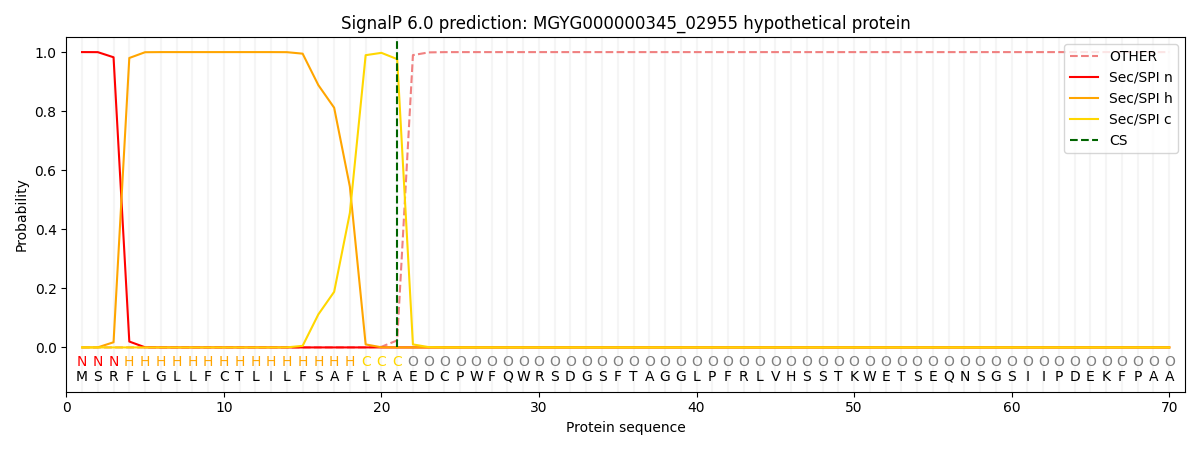
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000193
|
0.999226
|
0.000167
|
0.000137
|
0.000132
|
0.000126
|
There is no transmembrane helices in MGYG000000345_02955.

