You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000348_01817
You are here: Home > Sequence: MGYG000000348_01817
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; CAG-485; | |||||||||||
| CAZyme ID | MGYG000000348_01817 | |||||||||||
| CAZy Family | GH51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 16706; End: 21286 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH51 | 175 | 803 | 3.4e-104 | 0.8571428571428571 |
| GH43 | 969 | 1226 | 2.2e-43 | 0.9914163090128756 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd08983 | GH43_Bt3655-like | 2.29e-54 | 952 | 1240 | 2 | 262 | Glycosyl hydrolase family 43 protein such as Bacteroides thetaiotaomicron VPI-5482 arabinofuranosidase Bt3655. This glycosyl hydrolase family 43 (GH43)-like family includes the characterized arabinofuranosidases (EC 3.2.1.55): Bacteroides thetaiotaomicron VPI-5482 (Bt3655;BT_3655) and Penicillium chrysogenum 31B Abf43B, as well as Bifidobacterium adolescentis ATCC 15703 beta-xylosidase (EC 3.2.1.37) BAD_1527. It belongs to the glycosyl hydrolase clan F (according to carbohydrate-active enzymes database (CAZY)) which includes family 43 (GH43) and 62 (GH62) families. GH43 includes enzymes with beta-xylosidase (EC 3.2.1.37), beta-1,3-xylosidase (EC 3.2.1.-), alpha-L-arabinofuranosidase (EC 3.2.1.55), arabinanase (EC 3.2.1.99), xylanase (EC 3.2.1.8), endo-alpha-L-arabinanases (beta-xylanases) and galactan 1,3-beta-galactosidase (EC 3.2.1.145) activities. GH43 are inverting enzymes (i.e. they invert the stereochemistry of the anomeric carbon atom of the substrate) that have an aspartate as the catalytic general base, a glutamate as the catalytic general acid and another aspartate that is responsible for pKa modulation and orienting the catalytic acid. Many GH43 enzymes display both alpha-L-arabinofuranosidase and beta-D-xylosidase activity using aryl-glycosides as substrates. A common structural feature of GH43 enzymes is a 5-bladed beta-propeller domain that contains the catalytic acid and catalytic base. A long V-shaped groove, partially enclosed at one end, forms a single extended substrate-binding surface across the face of the propeller. |
| COG3534 | AbfA | 2.90e-42 | 200 | 797 | 34 | 492 | Alpha-L-arabinofuranosidase [Carbohydrate transport and metabolism]. |
| smart00813 | Alpha-L-AF_C | 6.00e-21 | 447 | 543 | 1 | 97 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| pfam06964 | Alpha-L-AF_C | 1.13e-20 | 447 | 539 | 1 | 96 | Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| pfam02018 | CBM_4_9 | 3.19e-07 | 59 | 195 | 1 | 134 | Carbohydrate binding domain. This family includes diverse carbohydrate binding domains. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGB28798.1 | 0.0 | 12 | 1517 | 11 | 1417 |
| QNT66893.1 | 2.04e-232 | 9 | 802 | 8 | 811 |
| BCS85815.1 | 1.59e-228 | 17 | 802 | 3 | 793 |
| ADE81175.1 | 1.91e-226 | 11 | 813 | 25 | 836 |
| AGB28822.1 | 3.40e-223 | 8 | 802 | 6 | 803 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ZPS_AAA | 2.15e-88 | 42 | 534 | 18 | 527 | ChainAAA, MgGH51 [Meripilus giganteus],6ZPV_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPW_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPX_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPY_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPZ_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ0_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ1_AAA Chain AAA, MgGH51 [Meripilus giganteus] |
| 4ATW_A | 9.09e-19 | 213 | 421 | 46 | 237 | Thecrystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_B The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_C The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_D The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_E The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8],4ATW_F The crystal structure of Arabinofuranosidase [Thermotoga maritima MSB8] |
| 3S2C_A | 9.22e-19 | 213 | 421 | 46 | 237 | Structureof the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_B Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_C Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_D Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_E Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_F Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_G Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_H Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_I Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_J Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_K Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3S2C_L Structure of the thermostable GH51 alpha-L-arabinofuranosidase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1] |
| 3UG3_A | 1.04e-18 | 213 | 421 | 66 | 257 | Crystalstructure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_B Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_C Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_D Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_E Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG3_F Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima ligand free form [Thermotoga maritima],3UG4_A Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_B Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_C Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_D Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_E Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG4_F Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima arabinose complex [Thermotoga maritima],3UG5_A Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_B Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_C Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_D Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_E Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima],3UG5_F Crystal structure of alpha-L-arabinofuranosidase from Thermotoga maritima xylose complex [Thermotoga maritima] |
| 2Y2W_A | 1.15e-15 | 213 | 360 | 89 | 215 | Elucidationof the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. [Bifidobacterium longum],2Y2W_B Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. [Bifidobacterium longum],2Y2W_C Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. [Bifidobacterium longum],2Y2W_D Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. [Bifidobacterium longum],2Y2W_E Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. [Bifidobacterium longum],2Y2W_F Elucidation of the substrate specificity and protein structure of AbfB, a family 51 alpha-L-arabinofuranosidase from Bifidobacterium longum. [Bifidobacterium longum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P82593 | 2.85e-140 | 11 | 802 | 20 | 821 | Extracellular exo-alpha-L-arabinofuranosidase OS=Streptomyces chartreusis OX=1969 PE=1 SV=1 |
| Q9SG80 | 9.56e-113 | 23 | 562 | 40 | 576 | Alpha-L-arabinofuranosidase 1 OS=Arabidopsis thaliana OX=3702 GN=ASD1 PE=1 SV=1 |
| Q8VZR2 | 2.33e-108 | 22 | 550 | 38 | 567 | Alpha-L-arabinofuranosidase 2 OS=Arabidopsis thaliana OX=3702 GN=ASD2 PE=2 SV=1 |
| Q8NK90 | 1.23e-85 | 3 | 527 | 11 | 528 | Alpha-L-arabinofuranosidase A OS=Aspergillus kawachii (strain NBRC 4308) OX=1033177 GN=abfA PE=1 SV=2 |
| Q96X54 | 2.27e-85 | 3 | 527 | 11 | 528 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus awamori OX=105351 GN=abfA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
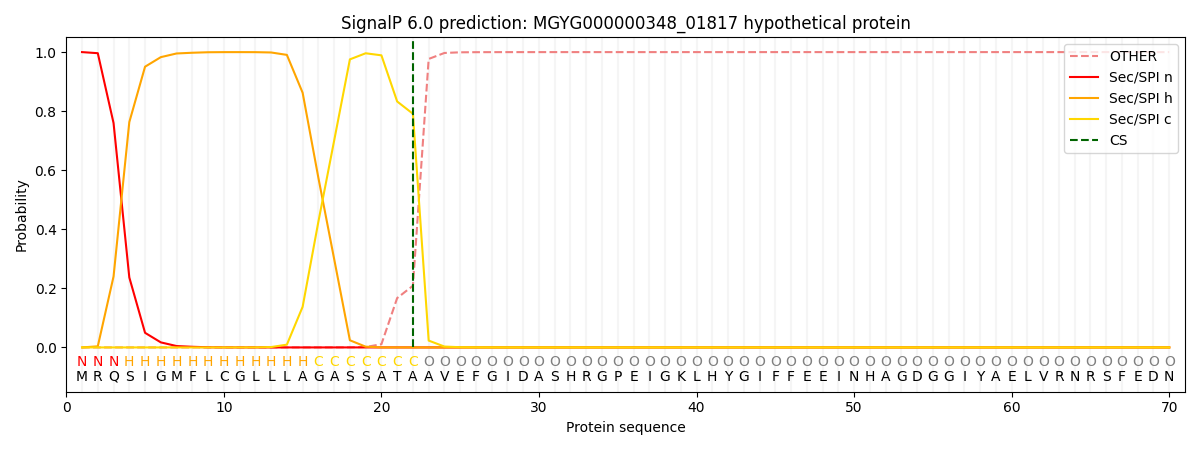
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000482 | 0.998697 | 0.000219 | 0.000213 | 0.000188 | 0.000182 |
