You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000355_00316
Basic Information
help
| Species |
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Tannerellaceae; Parabacteroides;
|
| CAZyme ID |
MGYG000000355_00316
|
| CAZy Family |
GT83 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 435 |
|
49483.97 |
9.4644 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000355 |
2806154 |
MAG |
Sweden |
Europe |
|
| Gene Location |
Start: 5074;
End: 6381
Strand: -
|
No EC number prediction in MGYG000000355_00316.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
17 |
428 |
6.5e-37 |
0.774074074074074 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1807
|
ArnT |
4.83e-14 |
18 |
422 |
13 |
418 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
C5BDQ8
|
3.95e-06 |
38 |
378 |
30 |
370 |
Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Edwardsiella ictaluri (strain 93-146) OX=634503 GN=arnT PE=3 SV=1 |
This protein is predicted as OTHER

| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.997373
|
0.002261
|
0.000088
|
0.000004
|
0.000003
|
0.000319
|
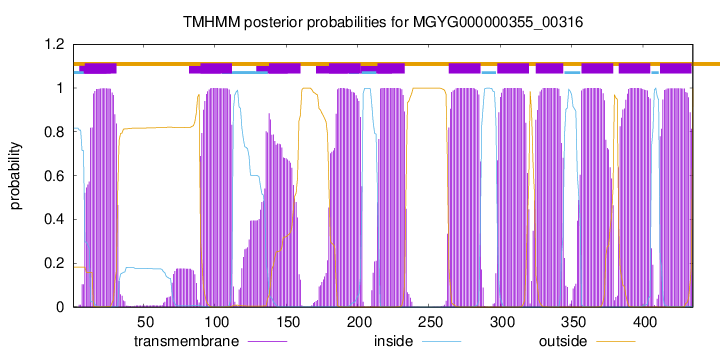
| start |
end |
| 9 |
31 |
| 90 |
112 |
| 138 |
160 |
| 180 |
202 |
| 214 |
233 |
| 264 |
286 |
| 298 |
320 |
| 325 |
344 |
| 357 |
379 |
| 383 |
405 |
| 412 |
434 |


