You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000356_02353
You are here: Home > Sequence: MGYG000000356_02353
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
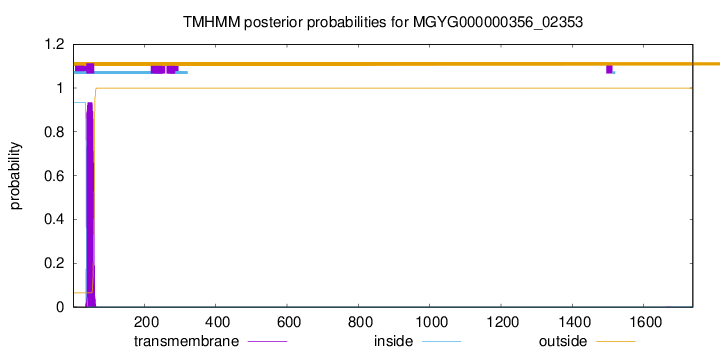
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; VUNI01; | |||||||||||
| CAZyme ID | MGYG000000356_02353 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12939; End: 18155 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 141 | 277 | 1.3e-17 | 0.9596774193548387 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 1.39e-18 | 139 | 277 | 1 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam09479 | Flg_new | 9.51e-13 | 1408 | 1471 | 2 | 65 | Listeria-Bacteroides repeat domain (List_Bact_rpt). This model describes a conserved core region of about 43 residues, which occurs in at least two families of tandem repeats. These include 78-residue repeats which occur from 2 to 15 times in some proteins of Bacteroides forsythus ATCC 43037, and 70-residue repeats found in families of internalins of Listeria species. Single copies are found in proteins of Fibrobacter succinogenes, Geobacter sulfurreducens, and a few other bacteria. |
| sd00036 | LRR_3 | 3.67e-11 | 1646 | 1704 | 24 | 81 | leucine-rich repeats. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
| cd00057 | FA58C | 7.79e-11 | 127 | 279 | 1 | 139 | Substituted updates: Jan 31, 2002 |
| sd00036 | LRR_3 | 9.54e-11 | 1646 | 1705 | 1 | 59 | leucine-rich repeats. A leucine-rich repeat (LRR) is a structural protein motif of 20-30 amino acids that is unusually rich in the hydrophobic amino acid leucine. The conserved eleven-residue sequence motif (LxxLxLxxN/CxL) within the LRRs corresponds to the beta-strand and adjacent loop regions, whereas the remaining parts of the repeats are variable. LRRs fold together to form a solenoid protein domain, termed leucine-rich repeat domain. Leucine-rich repeats are usually involved in protein-protein interactions. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT26415.1 | 3.08e-253 | 286 | 1160 | 86 | 914 |
| ALG48827.1 | 7.92e-251 | 32 | 1173 | 5 | 1182 |
| SQG39183.1 | 4.26e-250 | 32 | 1173 | 5 | 1182 |
| AMN35790.1 | 8.34e-250 | 32 | 1173 | 5 | 1182 |
| AXH52486.1 | 1.17e-249 | 32 | 1173 | 5 | 1182 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1K3I_A | 1.46e-14 | 112 | 287 | 8 | 170 | CrystalStructure of the Precursor of Galactose Oxidase [Fusarium sp.] |
| 2WQ8_A | 2.55e-14 | 119 | 287 | 17 | 175 | ChainA, GALACTOSE OXIDASE [Fusarium graminearum] |
| 2EIB_A | 5.61e-14 | 133 | 287 | 9 | 153 | ChainA, Galactose oxidase [Fusarium graminearum] |
| 1GOF_A | 5.61e-14 | 133 | 287 | 9 | 153 | NOVELTHIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],1GOG_A NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],1GOH_A NOVEL THIOETHER BOND REVEALED BY A 1.7 ANGSTROMS CRYSTAL STRUCTURE OF GALACTOSE OXIDASE [Hypomyces rosellus],2EIE_A Chain A, Galactose oxidase [Fusarium graminearum],2JKX_A Chain A, GALACTOSE OXIDASE [Fusarium graminearum],2VZ1_A Chain A, GALACTOSE OXIDASE [Fusarium graminearum],2VZ3_A Chain A, Galactose Oxidase [Fusarium graminearum] |
| 2EIC_A | 5.61e-14 | 133 | 287 | 9 | 153 | ChainA, Galactose oxidase [Fusarium graminearum] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| I1S2N3 | 6.33e-14 | 112 | 287 | 32 | 194 | Galactose oxidase OS=Gibberella zeae (strain ATCC MYA-4620 / CBS 123657 / FGSC 9075 / NRRL 31084 / PH-1) OX=229533 GN=GAOA PE=3 SV=1 |
| P0CS93 | 8.32e-14 | 112 | 287 | 32 | 194 | Galactose oxidase OS=Gibberella zeae OX=5518 GN=GAOA PE=1 SV=1 |
| P0DTR4 | 1.46e-10 | 133 | 281 | 509 | 645 | A type blood N-acetyl-alpha-D-galactosamine deacetylase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
| Q02834 | 2.61e-09 | 131 | 279 | 502 | 643 | Sialidase OS=Micromonospora viridifaciens OX=1881 GN=nedA PE=1 SV=1 |
| Q0TR53 | 3.27e-09 | 140 | 334 | 631 | 814 | O-GlcNAcase NagJ OS=Clostridium perfringens (strain ATCC 13124 / DSM 756 / JCM 1290 / NCIMB 6125 / NCTC 8237 / Type A) OX=195103 GN=nagJ PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
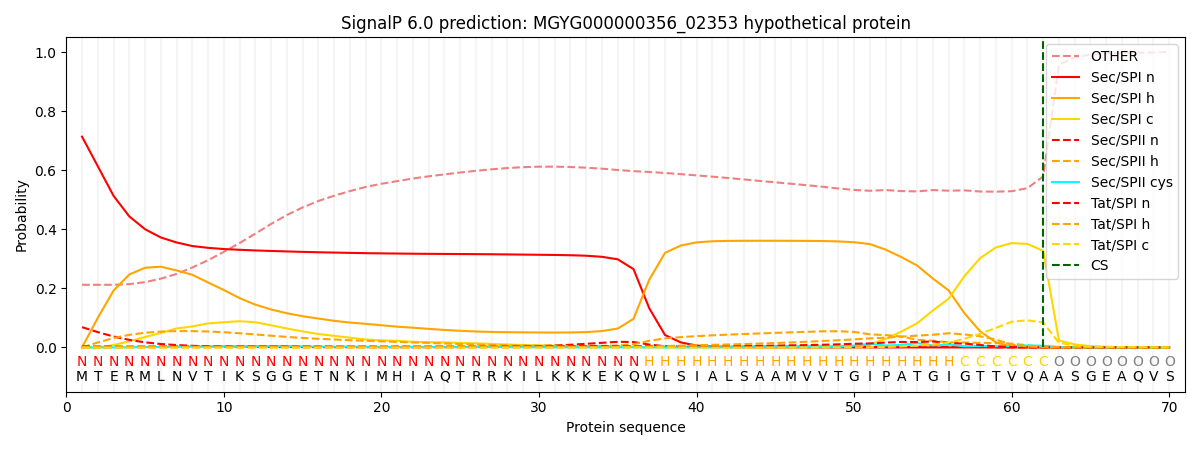
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.219994 | 0.622865 | 0.120606 | 0.032001 | 0.004085 | 0.000445 |