You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000360_00416
You are here: Home > Sequence: MGYG000000360_00416
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA1829 sp900548385 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA1829; UBA1829 sp900548385 | |||||||||||
| CAZyme ID | MGYG000000360_00416 | |||||||||||
| CAZy Family | GH148 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 540720; End: 544025 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH148 | 470 | 618 | 7.3e-39 | 0.9868421052631579 |
| GH148 | 681 | 797 | 1.5e-18 | 0.7763157894736842 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd18787 | SF2_C_DEAD | 0.008 | 484 | 551 | 36 | 89 | C-terminal helicase domain of the DEAD box helicases. DEAD-box helicases comprise a diverse family of proteins involved in ATP-dependent RNA unwinding, needed in a variety of cellular processes including splicing, ribosome biogenesis, and RNA degradation. They are superfamily (SF)2 helicases that, similar to SF1, do not form toroidal structures like SF3-6 helicases. Their helicase core consists of two similar protein domains that resemble the fold of the recombination protein RecA. This model describes the C-terminal domain, also called HelicC. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44571.1 | 0.0 | 28 | 1099 | 31 | 1114 |
| AVM44197.1 | 9.25e-296 | 1 | 1099 | 1 | 1094 |
| AVM43772.1 | 2.70e-257 | 123 | 1099 | 132 | 1108 |
| AVM44763.1 | 2.96e-251 | 121 | 1099 | 120 | 1112 |
| AHF90796.1 | 1.62e-194 | 101 | 1097 | 141 | 1174 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
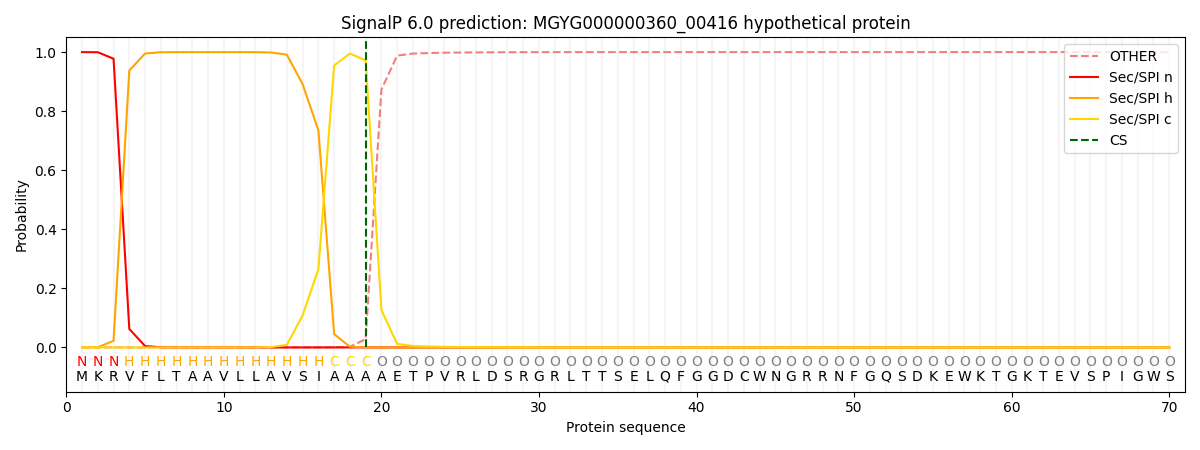
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000517 | 0.998550 | 0.000315 | 0.000211 | 0.000200 | 0.000187 |
