You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000360_00545
You are here: Home > Sequence: MGYG000000360_00545
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA1829 sp900548385 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA1829; UBA1829 sp900548385 | |||||||||||
| CAZyme ID | MGYG000000360_00545 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 714983; End: 716506 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 214 | 481 | 3.4e-45 | 0.7745454545454545 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 3.15e-24 | 219 | 467 | 27 | 271 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 7.29e-17 | 215 | 482 | 72 | 377 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam14488 | DUF4434 | 0.003 | 262 | 357 | 71 | 163 | Domain of unknown function (DUF4434). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44326.1 | 2.62e-189 | 2 | 507 | 12 | 519 |
| AVM44708.1 | 8.87e-122 | 3 | 506 | 2 | 499 |
| QNN21528.1 | 1.12e-94 | 1 | 507 | 9 | 519 |
| AVM46198.1 | 1.56e-91 | 95 | 506 | 78 | 501 |
| AVM45719.1 | 6.99e-90 | 14 | 504 | 63 | 549 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4LX4_A | 4.90e-16 | 258 | 459 | 79 | 275 | CrystalStructure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],4LX4_B Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],4LX4_C Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],4LX4_D Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],6R2J_A Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501],6R2J_B Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501],6R2J_C Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501],6R2J_D Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501] |
| 6KDD_A | 9.18e-14 | 292 | 469 | 116 | 295 | endoglucanase[Fervidobacterium pennivorans DSM 9078] |
| 7EC9_A | 1.08e-13 | 206 | 470 | 18 | 307 | ChainA, Endoglucanase [Thermotoga maritima MSB8],7EC9_B Chain B, Endoglucanase [Thermotoga maritima MSB8],7EFZ_A Chain A, Endoglucanase [Thermotoga maritima MSB8],7EFZ_B Chain B, Endoglucanase [Thermotoga maritima MSB8] |
| 1VJZ_A | 2.59e-13 | 206 | 470 | 18 | 307 | Crystalstructure of Endoglucanase (TM1752) from Thermotoga maritima at 2.05 A resolution [Thermotoga maritima] |
| 4EE9_A | 3.23e-09 | 229 | 438 | 42 | 247 | Crystalstructure of the RBcel1 endo-1,4-glucanase [uncultured bacterium],4M24_A Crystal structure of the endo-1,4-glucanase, RBcel1, in complex with cellobiose [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16169 | 8.00e-11 | 215 | 440 | 29 | 262 | Cellodextrinase A OS=Ruminococcus flavefaciens OX=1265 GN=celA PE=3 SV=3 |
| P25472 | 9.42e-10 | 270 | 463 | 110 | 298 | Endoglucanase D OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
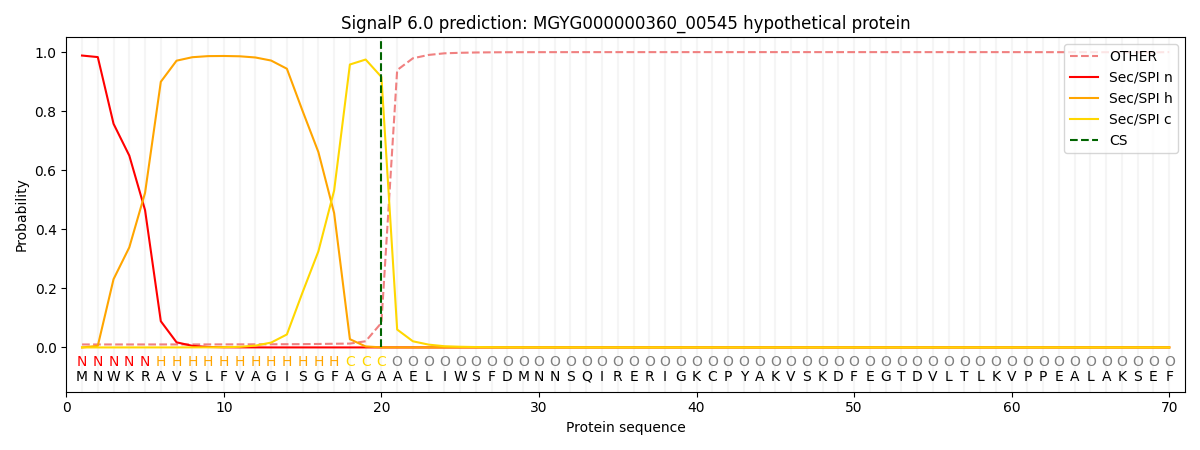
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.015346 | 0.981320 | 0.002354 | 0.000377 | 0.000294 | 0.000294 |

