You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000363_00699
You are here: Home > Sequence: MGYG000000363_00699
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Zag1 sp001917115 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Cyanobacteria; Vampirovibrionia; Gastranaerophilales; Gastranaerophilaceae; Zag1; Zag1 sp001917115 | |||||||||||
| CAZyme ID | MGYG000000363_00699 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 48270; End: 48908 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH23 | 42 | 150 | 7.5e-28 | 0.8222222222222222 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd00254 | LT-like | 4.92e-39 | 44 | 144 | 1 | 108 | lytic transglycosylase(LT)-like domain. Members include the soluble and insoluble membrane-bound LTs in bacteria and LTs in bacteriophage lambda. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| cd16896 | LT_Slt70-like | 5.52e-39 | 30 | 145 | 5 | 143 | uncharacterized lytic transglycosylase subfamily with similarity to Slt70. Uncharacterized lytic transglycosylase (LT) with a conserved sequence pattern suggesting similarity to the Slt70, a 70kda soluble lytic transglycosylase which also has an N-terminal U-shaped U-domain and a linker L-domain. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| cd13401 | Slt70-like | 2.05e-37 | 30 | 144 | 7 | 144 | 70kDa soluble lytic transglycosylase (Slt70) and similar proteins. Catalytic domain of the 70kda soluble lytic transglycosylase (LT)-like proteins, which also have an N-terminal U-shaped U-domain and a linker L-domain. LTs catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc), as do "goose-type" lysozymes. However, in addition to this, they also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria and the LTs in bacteriophage lambda. |
| cd13403 | MLTF-like | 9.26e-27 | 33 | 144 | 1 | 154 | membrane-bound lytic murein transglycosylase F (MLTF) and similar proteins. This subfamily includes membrane-bound lytic murein transglycosylase F (MltF, murein lyase F) that degrades murein glycan strands. It is responsible for catalyzing the release of 1,6-anhydromuropeptides from peptidoglycan. Lytic transglycosylase catalyzes the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc) as do goose-type lysozymes. However, in addition, it also makes a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. |
| cd16894 | MltD-like | 2.36e-26 | 54 | 143 | 17 | 125 | Membrane-bound lytic murein transglycosylase D and similar proteins. Lytic transglycosylases (LT) catalyze the cleavage of the beta-1,4-glycosidic bond between N-acetylmuramic acid (MurNAc) and N-acetyl-D-glucosamine (GlcNAc). Membrane-bound lytic murein transglycosylase D protein (MltD) family members may have one or more small LysM domains, which may contribute to peptidoglycan binding. Unlike the similar "goose-type" lysozymes, LTs also make a new glycosidic bond with the C6 hydroxyl group of the same muramic acid residue. Proteins similar to this family include the soluble and insoluble membrane-bound LTs in bacteria, the LTs in bacteriophage lambda, as well as the eukaryotic "goose-type" lysozymes (goose egg-white lysozyme; GEWL). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AOR39353.1 | 5.67e-116 | 1 | 212 | 1 | 216 |
| ADI02565.1 | 1.09e-32 | 14 | 148 | 79 | 212 |
| AZT90545.1 | 2.93e-31 | 35 | 143 | 101 | 210 |
| AQS60617.1 | 3.20e-31 | 22 | 144 | 79 | 203 |
| AIF43250.1 | 1.14e-30 | 27 | 143 | 85 | 203 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6FBT_A | 8.84e-15 | 30 | 165 | 450 | 610 | ChainA, Lytic murein transglycosylase [Pseudomonas aeruginosa] |
| 5OHU_A | 8.97e-15 | 30 | 165 | 479 | 639 | TheX-ray Structure of Lytic Transglycosylase Slt from Pseudomonas aeruginosa [Pseudomonas aeruginosa] |
| 6FC4_A | 2.20e-14 | 30 | 165 | 451 | 611 | ChainA, Soluble lytic murein transglycosylase [Pseudomonas aeruginosa] |
| 6FCQ_A | 2.20e-14 | 30 | 165 | 450 | 610 | ChainA, Soluble lytic murein transglycosylase [Pseudomonas aeruginosa],6FCR_A Chain A, Soluble lytic murein transglycosylase [Pseudomonas aeruginosa],6FCS_A Chain A, Soluble lytic murein transglycosylase [Pseudomonas aeruginosa],6FCU_A Chain A, Soluble lytic murein transglycosylase [Pseudomonas aeruginosa] |
| 5MPQ_A | 2.93e-14 | 26 | 145 | 413 | 551 | BulgecinA: The key to a broad-spectrum inhibitor that targets lytic transglycosylases [Neisseria meningitidis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O31608 | 6.15e-28 | 28 | 144 | 59 | 177 | Putative murein lytic transglycosylase YjbJ OS=Bacillus subtilis (strain 168) OX=224308 GN=yjbJ PE=3 SV=1 |
| O31976 | 2.56e-26 | 37 | 168 | 1430 | 1559 | SPbeta prophage-derived uncharacterized transglycosylase YomI OS=Bacillus subtilis (strain 168) OX=224308 GN=yomI PE=3 SV=2 |
| O64046 | 2.56e-26 | 37 | 168 | 1430 | 1559 | Probable tape measure protein OS=Bacillus phage SPbeta OX=66797 GN=yomI PE=3 SV=1 |
| P27380 | 6.49e-12 | 32 | 160 | 10 | 133 | Transglycosylase OS=Enterobacteria phage PRD1 OX=10658 GN=VII PE=1 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
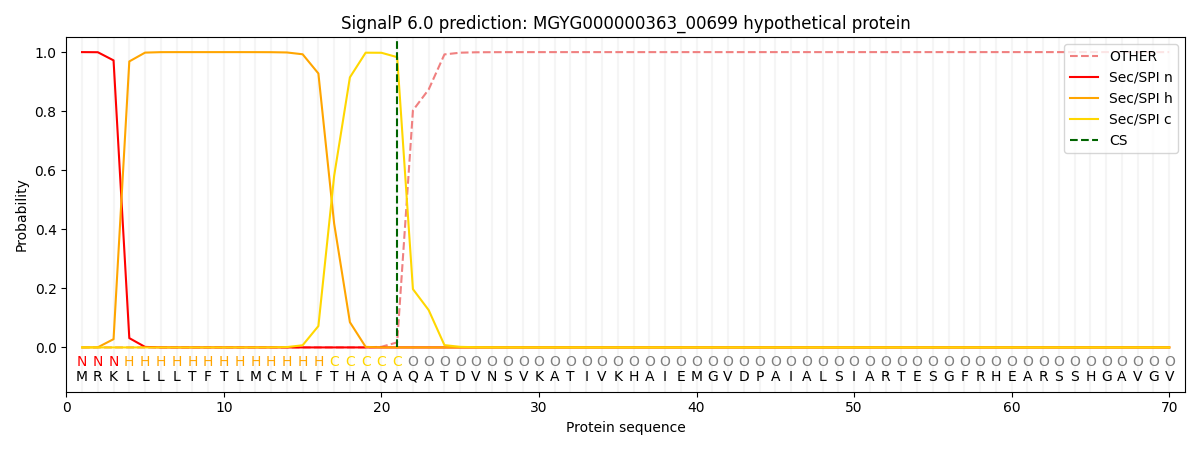
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000219 | 0.999107 | 0.000179 | 0.000173 | 0.000153 | 0.000141 |
