You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000364_01181
You are here: Home > Sequence: MGYG000000364_01181
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
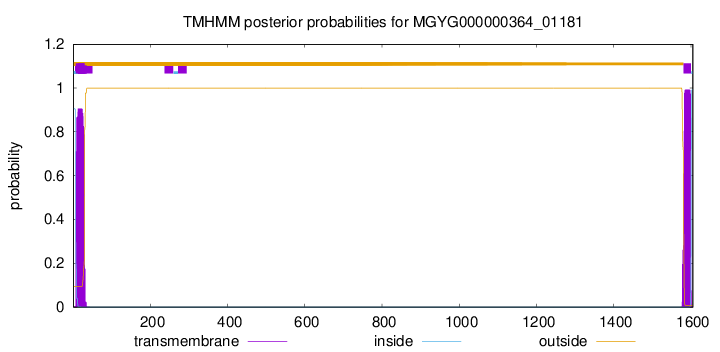
TMHMM annotations
Basic Information help
| Species | Coprococcus sp900767685 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Coprococcus; Coprococcus sp900767685 | |||||||||||
| CAZyme ID | MGYG000000364_01181 | |||||||||||
| CAZy Family | CBM27 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 49066; End: 53883 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 779 | 1116 | 2.3e-62 | 0.900990099009901 |
| CBM23 | 1286 | 1447 | 1.5e-45 | 0.9691358024691358 |
| CBM27 | 595 | 745 | 1.6e-31 | 0.9821428571428571 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02156 | Glyco_hydro_26 | 1.36e-38 | 803 | 1060 | 24 | 264 | Glycosyl hydrolase family 26. |
| COG4124 | ManB2 | 4.31e-19 | 849 | 1126 | 91 | 352 | Beta-mannanase [Carbohydrate transport and metabolism]. |
| pfam03425 | CBM_11 | 1.58e-06 | 1287 | 1429 | 7 | 155 | Carbohydrate binding domain (family 11). |
| PRK09418 | PRK09418 | 2.05e-04 | 1466 | 1605 | 642 | 777 | bifunctional 2',3'-cyclic-nucleotide 2'-phosphodiesterase/3'-nucleotidase. |
| pfam09212 | CBM27 | 4.52e-04 | 583 | 745 | 9 | 170 | Carbohydrate binding module 27. Members of this family are carbohydrate binding modules that bind to beta-1, 4-manno-oligosaccharides, carob galactomannan, and konjac glucomannan, but not to cellulose (insoluble and soluble) or soluble birchwood xylan. They adopt a beta sandwich structure comprising 13 beta strands with a single, small alpha-helix and a single metal atom. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBK83362.1 | 0.0 | 1 | 1605 | 1 | 1544 |
| VCV20359.1 | 1.81e-294 | 424 | 1482 | 34 | 1074 |
| CBL14297.1 | 1.81e-294 | 424 | 1482 | 34 | 1074 |
| CBL07467.1 | 2.18e-294 | 424 | 1482 | 34 | 1074 |
| EEV00397.1 | 2.61e-294 | 424 | 1482 | 46 | 1086 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4YN5_A | 5.99e-52 | 775 | 1152 | 48 | 414 | Catalyticdomain of Bacillus sp. JAMB-750 GH26 Endo-beta-1,4-mannanase [Bacillus sp. JAMB750] |
| 1J9Y_A | 2.22e-42 | 773 | 1155 | 6 | 372 | Crystalstructure of mannanase 26A from Pseudomonas cellulosa [Cellvibrio japonicus] |
| 1R7O_A | 2.79e-42 | 773 | 1155 | 16 | 382 | CrystalStructure of apo-mannanase 26A from Psudomonas cellulosa [Cellvibrio japonicus] |
| 2WHM_A | 1.01e-41 | 773 | 1155 | 6 | 372 | Cellvibriojaponicus Man26A E121A and E320G double mutant in complex with mannobiose [Cellvibrio japonicus] |
| 1GW1_A | 1.12e-41 | 773 | 1155 | 2 | 365 | Substratedistortion by beta-mannanase from Pseudomonas cellulosa [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A1A278 | 5.74e-50 | 774 | 1422 | 38 | 653 | Mannan endo-1,4-beta-mannosidase OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=BAD_1030 PE=1 SV=1 |
| P49424 | 2.83e-41 | 773 | 1155 | 44 | 410 | Mannan endo-1,4-beta-mannosidase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=manA PE=1 SV=2 |
| O05512 | 6.09e-14 | 782 | 1024 | 35 | 268 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis (strain 168) OX=224308 GN=gmuG PE=1 SV=2 |
| Q5AWB7 | 1.02e-13 | 930 | 1075 | 181 | 311 | Probable mannan endo-1,4-beta-mannosidase E OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=manE PE=3 SV=1 |
| P55278 | 1.07e-13 | 908 | 1024 | 144 | 266 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis OX=1423 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
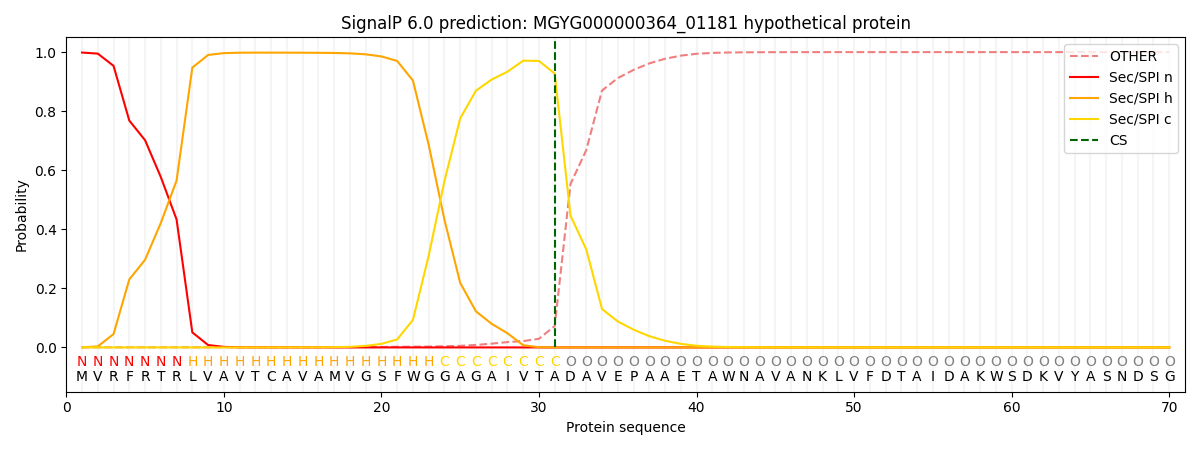
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001066 | 0.996569 | 0.001290 | 0.000634 | 0.000219 | 0.000186 |