You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000367_00685
You are here: Home > Sequence: MGYG000000367_00685
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
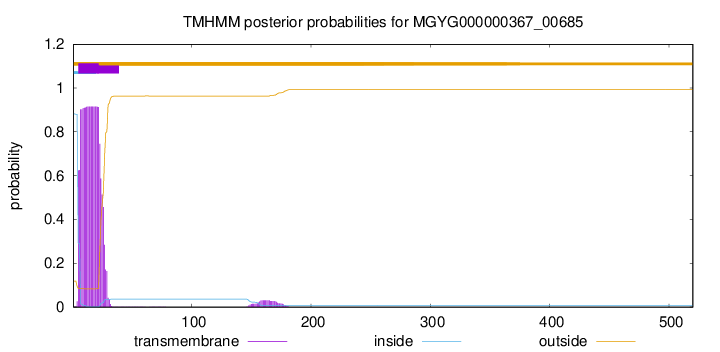
TMHMM annotations
Basic Information help
| Species | QAKD01 sp003343965 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Peptostreptococcales; Anaerovoracaceae; QAKD01; QAKD01 sp003343965 | |||||||||||
| CAZyme ID | MGYG000000367_00685 | |||||||||||
| CAZy Family | GH26 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 27568; End: 29130 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH26 | 303 | 426 | 2.2e-29 | 0.37293729372937295 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02156 | Glyco_hydro_26 | 1.72e-14 | 300 | 415 | 129 | 242 | Glycosyl hydrolase family 26. |
| COG4124 | ManB2 | 7.57e-14 | 302 | 481 | 155 | 340 | Beta-mannanase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QHI71412.1 | 2.57e-219 | 9 | 520 | 4 | 517 |
| QAT42469.1 | 1.16e-218 | 9 | 520 | 7 | 520 |
| QIB68979.1 | 9.19e-215 | 1 | 519 | 1 | 516 |
| QOX62356.1 | 4.19e-197 | 29 | 519 | 26 | 520 |
| QUH19438.1 | 1.00e-189 | 41 | 511 | 50 | 524 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2VI0_A | 9.31e-31 | 279 | 510 | 59 | 280 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2BVD_A | 1.31e-30 | 279 | 510 | 59 | 280 | ChainA, ENDOGLUCANASE H [Acetivibrio thermocellus] |
| 2V3G_A | 1.31e-30 | 279 | 510 | 59 | 280 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2BV9_A | 1.53e-30 | 279 | 510 | 59 | 280 | ChainA, Endoglucanase H [Acetivibrio thermocellus] |
| 2CIP_A | 3.32e-30 | 279 | 510 | 59 | 280 | ChainA, ENDOGLUCANASE H [Acetivibrio thermocellus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16218 | 8.00e-28 | 279 | 510 | 81 | 302 | Endoglucanase H OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celH PE=1 SV=1 |
| P55278 | 5.48e-10 | 316 | 414 | 175 | 271 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis OX=1423 PE=3 SV=1 |
| O05512 | 1.31e-09 | 283 | 414 | 153 | 273 | Mannan endo-1,4-beta-mannosidase OS=Bacillus subtilis (strain 168) OX=224308 GN=gmuG PE=1 SV=2 |
| P16699 | 1.81e-07 | 279 | 410 | 138 | 271 | Mannan endo-1,4-beta-mannosidase A and B OS=Caldalkalibacillus mannanilyticus (strain DSM 16130 / CIP 109019 / JCM 10596 / AM-001) OX=1236954 PE=1 SV=1 |
| A1A278 | 2.22e-06 | 265 | 411 | 119 | 280 | Mannan endo-1,4-beta-mannosidase OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=BAD_1030 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
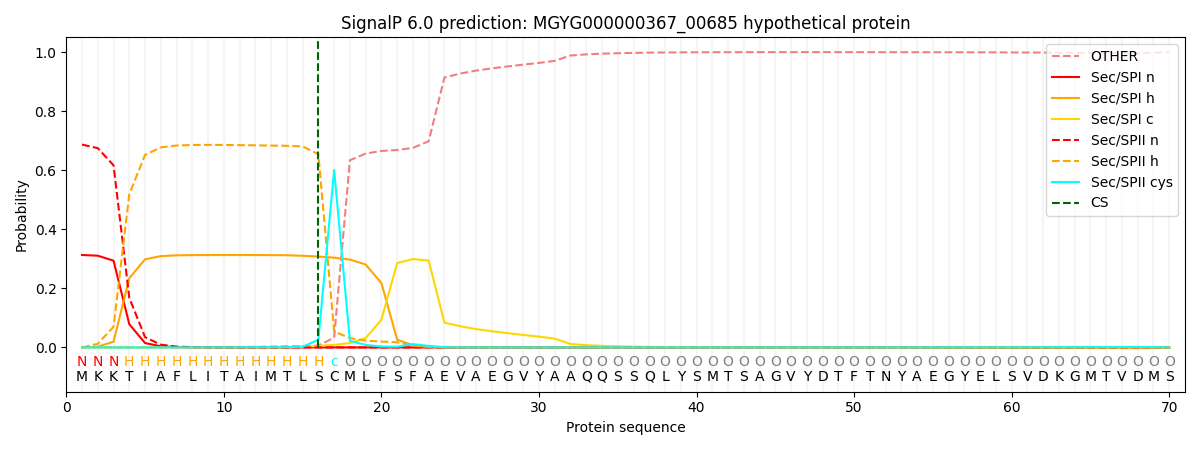
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001162 | 0.302758 | 0.694912 | 0.000550 | 0.000356 | 0.000262 |