You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000380_00802
You are here: Home > Sequence: MGYG000000380_00802
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
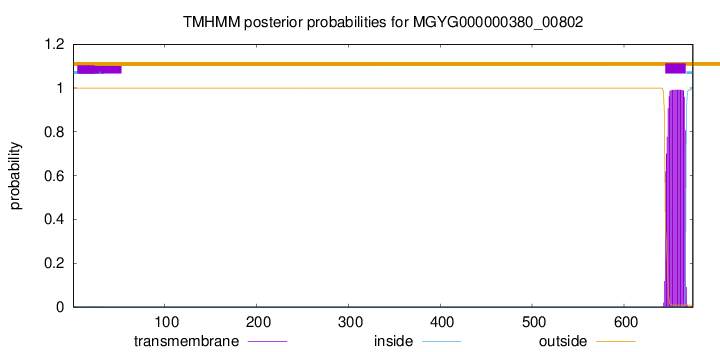
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelotrichaceae; Merdibacter; | |||||||||||
| CAZyme ID | MGYG000000380_00802 | |||||||||||
| CAZy Family | GH51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 164; End: 2191 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00813 | Alpha-L-AF_C | 8.49e-14 | 335 | 456 | 65 | 189 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| pfam06964 | Alpha-L-AF_C | 5.97e-13 | 335 | 456 | 68 | 192 | Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| COG3534 | AbfA | 7.09e-11 | 314 | 465 | 353 | 501 | Alpha-L-arabinofuranosidase [Carbohydrate transport and metabolism]. |
| TIGR02243 | TIGR02243 | 1.15e-08 | 86 | 150 | 373 | 442 | putative baseplate assembly protein. This family consists of a large, conserved hypothetical protein in phage tail-like regions of at least six bacterial genomes: Gloeobacter violaceus PCC 7421, Geobacter sulfurreducens PCA, Streptomyces coelicolor A3(2), Streptomyces avermitilis MA-4680, Mesorhizobium loti, and Myxococcus xanthus. The C-terminal region is identified by the broader model pfam04865 as related to baseplate protein J from phage P2, but that relationship is not observed directly. [Mobile and extrachromosomal element functions, Prophage functions] |
| pfam16403 | DUF5011 | 7.89e-07 | 484 | 549 | 10 | 71 | Domain of unknown function (DUF5011). This small family of proteins is functionally uncharacterized. This family is found in Bacteroides, Prevotella, and Parabateroides. Proteins in this family are around 230 amino acids in length. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ALE10461.1 | 1.40e-219 | 7 | 552 | 510 | 1060 |
| BBA48000.1 | 5.55e-219 | 7 | 552 | 510 | 1060 |
| AXM90856.1 | 6.33e-219 | 7 | 552 | 527 | 1077 |
| AFL03619.1 | 7.83e-219 | 7 | 552 | 510 | 1060 |
| BBA56144.1 | 7.83e-219 | 7 | 552 | 510 | 1060 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2KPN_A | 6.75e-09 | 484 | 552 | 20 | 88 | SolutionNMR structure of a Bacterial Ig-like (Big_3) domain from Bacillus cereus. Northeast Structural Genomics Consortium target BcR147A [Bacillus cereus ATCC 14579] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P56957 | 3.34e-07 | 476 | 559 | 261 | 347 | Pesticidal crystal protein Cry22Aa OS=Bacillus thuringiensis OX=1428 GN=cry22Aa PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
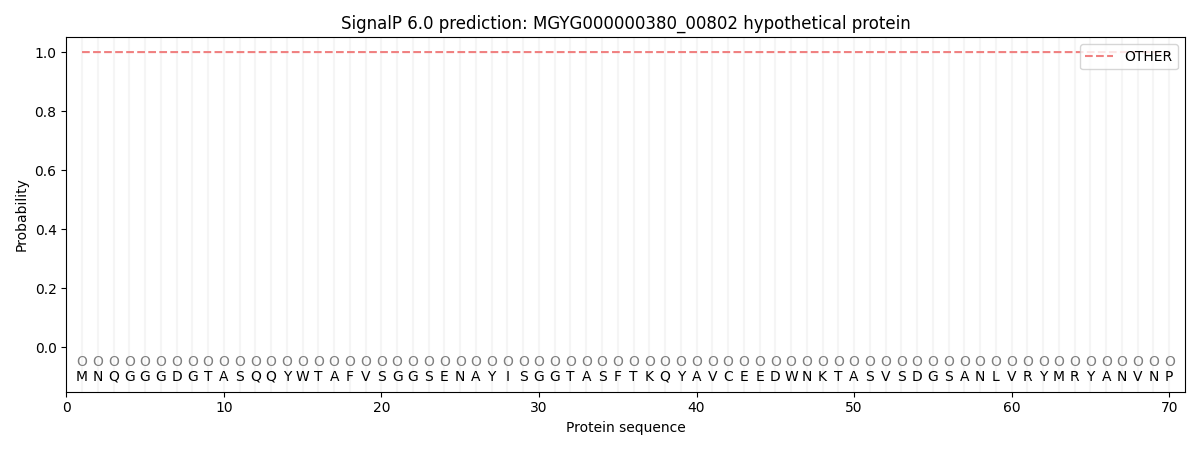
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000062 | 0.000002 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |