You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000387_01936
You are here: Home > Sequence: MGYG000000387_01936
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; ; | |||||||||||
| CAZyme ID | MGYG000000387_01936 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1758; End: 4523 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 167 | 436 | 1.9e-32 | 0.8465346534653465 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3866 | PelB | 2.21e-11 | 167 | 389 | 93 | 249 | Pectate lyase [Carbohydrate transport and metabolism]. |
| cd14256 | Dockerin_I | 3.01e-09 | 862 | 918 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam00404 | Dockerin_1 | 2.18e-06 | 863 | 919 | 1 | 56 | Dockerin type I repeat. The dockerin repeat is the binding partner of the cohesin domain pfam00963. The cohesin-dockerin interaction is the crucial interaction for complex formation in the cellulosome. The dockerin repeats, each bearing homology to the EF-hand calcium-binding loop bind calcium. |
| smart00656 | Amb_all | 8.07e-06 | 192 | 391 | 34 | 166 | Amb_all domain. |
| pfam02368 | Big_2 | 9.48e-06 | 598 | 664 | 3 | 64 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGH35942.1 | 7.14e-132 | 54 | 576 | 36 | 548 |
| QNF29855.1 | 1.98e-131 | 54 | 581 | 36 | 553 |
| ADL50775.1 | 1.17e-129 | 52 | 609 | 35 | 581 |
| BAV13078.1 | 1.27e-129 | 52 | 609 | 38 | 584 |
| AFH62972.1 | 3.15e-126 | 54 | 578 | 35 | 550 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5GT5_A | 1.95e-18 | 88 | 433 | 27 | 341 | Structuralbasis of the specific activity and thermostability of pectate lyase (pelN) from Paenibacillus sp. 0602 [Paenibacillus sp. 0602],5GT5_B Structural basis of the specific activity and thermostability of pectate lyase (pelN) from Paenibacillus sp. 0602 [Paenibacillus sp. 0602] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D3JTC2 | 8.99e-17 | 88 | 449 | 56 | 385 | Pectate lyase B OS=Paenibacillus amylolyticus OX=1451 GN=pelB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
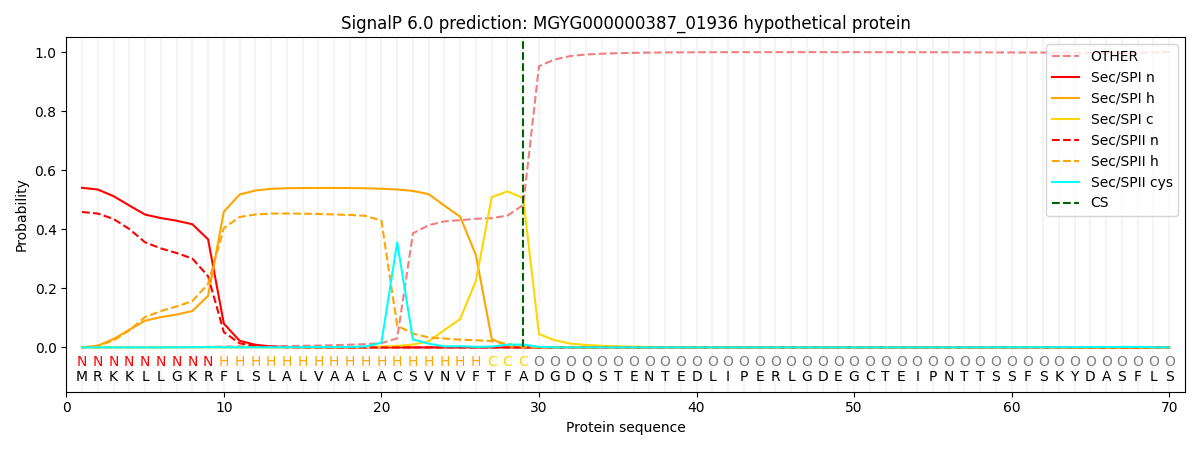
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001390 | 0.531687 | 0.466039 | 0.000434 | 0.000239 | 0.000189 |
