You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000393_00670
You are here: Home > Sequence: MGYG000000393_00670
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; | |||||||||||
| CAZyme ID | MGYG000000393_00670 | |||||||||||
| CAZy Family | GH163 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5427; End: 7673 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH163 | 211 | 465 | 1.3e-107 | 0.9960159362549801 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam16126 | DUF4838 | 4.89e-137 | 204 | 465 | 1 | 263 | Domain of unknown function (DUF4838). This family consists of several uncharacterized proteins found in various Bacteroides and Chloroflexus species. The function of this family is unknown. |
| pfam00754 | F5_F8_type_C | 1.65e-11 | 616 | 726 | 16 | 117 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam02838 | Glyco_hydro_20b | 3.24e-05 | 30 | 135 | 21 | 105 | Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold. |
| cd00057 | FA58C | 0.001 | 630 | 738 | 36 | 136 | Substituted updates: Jan 31, 2002 |
| cd14791 | GH36 | 0.002 | 213 | 315 | 92 | 194 | glycosyl hydrolase family 36 (GH36). GH36 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-galactosidase, alpha-N-acetylgalactosaminidase, stachyose synthase, and raffinose synthase. All GH36 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH36 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCA50059.1 | 0.0 | 10 | 747 | 18 | 740 |
| QUT70943.1 | 0.0 | 10 | 747 | 10 | 732 |
| AAO76142.1 | 0.0 | 20 | 747 | 24 | 733 |
| QMW88052.1 | 0.0 | 20 | 747 | 24 | 733 |
| ALJ42190.1 | 0.0 | 20 | 747 | 24 | 733 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6Q63_A | 2.23e-18 | 583 | 747 | 607 | 773 | BT0459[Bacteroides thetaiotaomicron],6Q63_B BT0459 [Bacteroides thetaiotaomicron],6Q63_C BT0459 [Bacteroides thetaiotaomicron] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
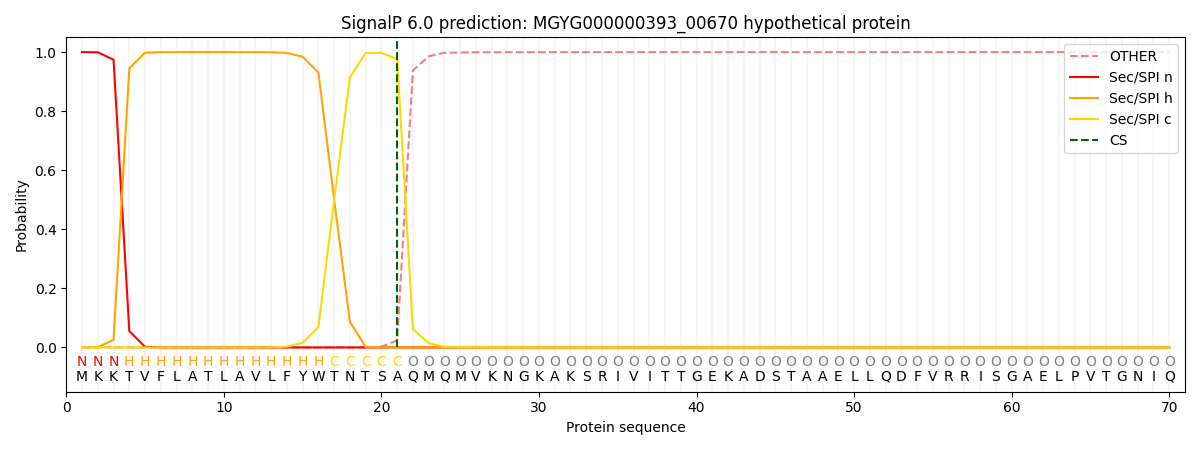
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000352 | 0.998919 | 0.000219 | 0.000159 | 0.000166 | 0.000164 |
