You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000393_01032
You are here: Home > Sequence: MGYG000000393_01032
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; | |||||||||||
| CAZyme ID | MGYG000000393_01032 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1088; End: 2479 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 61 | 419 | 3.7e-39 | 0.8352668213457076 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01229 | Glyco_hydro_39 | 5.41e-17 | 58 | 368 | 42 | 349 | Glycosyl hydrolases family 39. |
| cd21510 | agarase_cat | 2.66e-04 | 170 | 256 | 100 | 187 | alpha-beta barrel catalytic domain of agarase, such as GH86-like endo-acting agarases identified in non-marine organisms. Typically, agarases (E.C. 3.2.1.81) are found in ocean-dwelling bacteria since agarose is a principle component of red algae cell wall polysaccharides. Agarose is a linear polymer of alternating D-galactose and 3,6-anhydro-L-galactopyranose. Endo-acting agarases, such as glycoside hydrolase 16 (GH16) and GH86 hydrolyze internal beta-1,4 linkages. GH86-like endo-acting agarase of this protein family has been identified in the human intestinal bacterium Bacteroides uniformis. This acquired metabolic pathway, as demonstrated by the prevalence of agar-specific genetic cluster called polysaccharide utilization loci (PULs), varies considerably between human populations, being much more prevalent in a Japanese sample than in North America, European, or Chinese samples. Agarase activity was also identified in the non-marine bacterium Cellvibrio sp. |
| cd06542 | GH18_EndoS-like | 0.008 | 82 | 179 | 28 | 122 | Endo-beta-N-acetylglucosaminidases are bacterial chitinases that hydrolyze the chitin core of various asparagine (N)-linked glycans and glycoproteins. The endo-beta-N-acetylglucosaminidases have a glycosyl hydrolase family 18 (GH18) catalytic domain. Some members also have an additional C-terminal glycosyl hydrolase family 20 (GH20) domain while others have an N-terminal domain of unknown function (pfam08522). Members of this family include endo-beta-N-acetylglucosaminidase S (EndoS) from Streptococcus pyogenes, EndoF1, EndoF2, EndoF3, and EndoH from Flavobacterium meningosepticum, and EndoE from Enterococcus faecalis. EndoS is a secreted endoglycosidase from Streptococcus pyogenes that specifically hydrolyzes the glycan on human IgG between two core N-acetylglucosamine residues. EndoE is a secreted endoglycosidase, encoded by the ndoE gene in Enterococcus faecalis, that hydrolyzes the glycan on human RNase B. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QJD82796.1 | 2.46e-72 | 23 | 459 | 3 | 436 |
| QTH43246.1 | 6.66e-72 | 32 | 401 | 12 | 379 |
| QJD85864.1 | 7.82e-72 | 21 | 456 | 2 | 441 |
| AVM69820.1 | 1.67e-71 | 23 | 441 | 3 | 426 |
| AVM46100.1 | 1.67e-68 | 26 | 408 | 7 | 384 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5Z3K_A | 1.09e-33 | 77 | 342 | 58 | 311 | Crystalstructure of glucosidase from Croceicoccus marinus at 1.8 Angstrom resolution [Croceicoccus marinus],5Z3K_B Crystal structure of glucosidase from Croceicoccus marinus at 1.8 Angstrom resolution [Croceicoccus marinus] |
| 6YYH_A | 2.03e-14 | 101 | 258 | 100 | 255 | Crystalstructure of beta-D-xylosidase from Dictyoglomus thermophilum in ligand-free form [Dictyoglomus thermophilum H-6-12],6YYH_B Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum in ligand-free form [Dictyoglomus thermophilum H-6-12],6YYI_A Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum bound to beta-D-xylopyranose [Dictyoglomus thermophilum H-6-12],6YYI_B Crystal structure of beta-D-xylosidase from Dictyoglomus thermophilum bound to beta-D-xylopyranose [Dictyoglomus thermophilum H-6-12] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
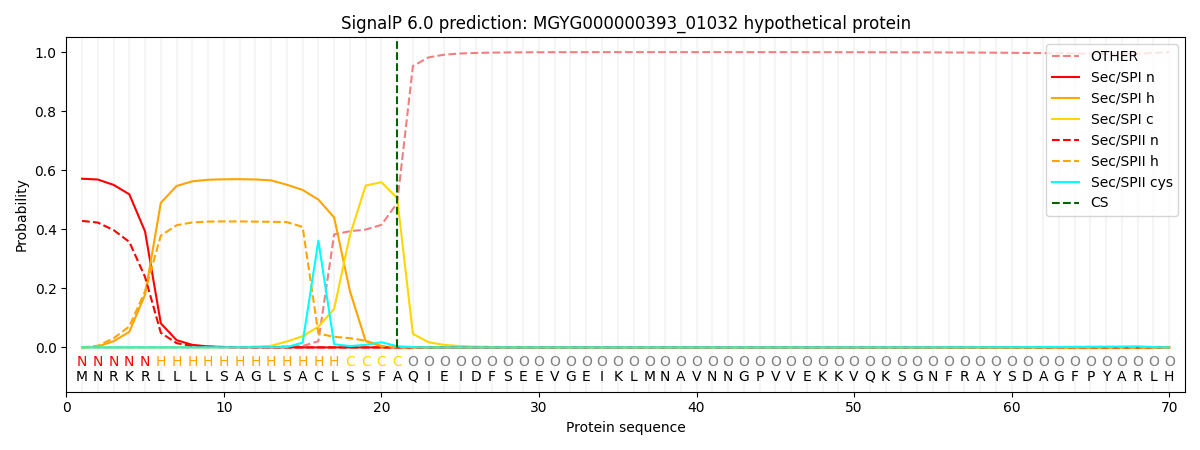
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001493 | 0.555742 | 0.441058 | 0.000691 | 0.000578 | 0.000415 |
