You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000398_01846
You are here: Home > Sequence: MGYG000000398_01846
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; TF01-11; | |||||||||||
| CAZyme ID | MGYG000000398_01846 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 102647; End: 105502 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 68 | 371 | 2.8e-100 | 0.9927536231884058 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 6.99e-61 | 65 | 372 | 12 | 270 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 1.02e-30 | 41 | 336 | 43 | 326 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam02389 | Cornifin | 7.83e-13 | 651 | 746 | 30 | 128 | Cornifin (SPRR) family. SPRR genes (formerly SPR) encode a novel class of polypeptides (small proline rich proteins) that are strongly induced during differentiation of human epidermal keratinocytes in vitro and in vivo. The most characteristic feature of the SPRR gene family resides in the structure of the central segments of the encoded polypeptides that are built up from tandemly repeated units of either eight (SPRR1 and SPRR3) or nine (SPRR2) amino acids with the general consensus XKXPEPXX where X is any amino acid. In order to avoid bacterial contamination due to the high polar-nature of the HMM the threshold has been set very high. |
| pfam02389 | Cornifin | 5.69e-12 | 647 | 761 | 18 | 131 | Cornifin (SPRR) family. SPRR genes (formerly SPR) encode a novel class of polypeptides (small proline rich proteins) that are strongly induced during differentiation of human epidermal keratinocytes in vitro and in vivo. The most characteristic feature of the SPRR gene family resides in the structure of the central segments of the encoded polypeptides that are built up from tandemly repeated units of either eight (SPRR1 and SPRR3) or nine (SPRR2) amino acids with the general consensus XKXPEPXX where X is any amino acid. In order to avoid bacterial contamination due to the high polar-nature of the HMM the threshold has been set very high. |
| pfam05109 | Herpes_BLLF1 | 1.47e-10 | 645 | 824 | 486 | 663 | Herpes virus major outer envelope glycoprotein (BLLF1). This family consists of the BLLF1 viral late glycoprotein, also termed gp350/220. It is the most abundantly expressed glycoprotein in the viral envelope of the Herpesviruses and is the major antigen responsible for stimulating the production of neutralising antibodies in vivo. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCN29385.1 | 8.52e-198 | 18 | 655 | 330 | 965 |
| QAA35398.1 | 4.17e-160 | 35 | 398 | 37 | 400 |
| ADL50682.1 | 6.78e-158 | 1 | 398 | 1 | 399 |
| BAV13070.1 | 8.36e-158 | 1 | 398 | 7 | 405 |
| AAR65335.1 | 5.50e-157 | 41 | 399 | 34 | 392 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2JEP_A | 2.01e-158 | 41 | 399 | 34 | 392 | Nativefamily 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEP_B Native family 5 xyloglucanase from Paenibacillus pabuli [Paenibacillus pabuli],2JEQ_A Family 5 xyloglucanase from Paenibacillus pabuli in complex with ligand [Paenibacillus pabuli] |
| 6WQY_A | 1.56e-69 | 33 | 399 | 12 | 380 | ChainA, Cellulase [Phocaeicola salanitronis DSM 18170] |
| 4W8A_A | 5.63e-63 | 41 | 374 | 2 | 339 | Crystalstructure of XEG5B, a GH5 xyloglucan-specific beta-1,4-glucanase from ruminal metagenomic library, in the native form [uncultured bacterium],4W8B_A Crystal structure of XEG5B, a GH5 xyloglucan-specific beta-1,4-glucanase from ruminal metagenomic library, in complex with XXLG [uncultured bacterium] |
| 4IM4_A | 1.12e-60 | 36 | 399 | 1 | 333 | ChainA, Endoglucanase E [Acetivibrio thermocellus],4IM4_B Chain B, Endoglucanase E [Acetivibrio thermocellus],4IM4_C Chain C, Endoglucanase E [Acetivibrio thermocellus],4IM4_D Chain D, Endoglucanase E [Acetivibrio thermocellus],4IM4_E Chain E, Endoglucanase E [Acetivibrio thermocellus],4IM4_F Chain F, Endoglucanase E [Acetivibrio thermocellus] |
| 6WQP_A | 2.34e-59 | 37 | 399 | 11 | 354 | GH5-4broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQP_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis [Ruminococcus champanellensis],6WQV_A GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_B GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_C GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis],6WQV_D GH5-4 broad specificity endoglucanase from Ruminococcus champanellensis with bound cellotriose [Ruminococcus champanellensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O08342 | 3.29e-148 | 28 | 399 | 28 | 397 | Endoglucanase A OS=Paenibacillus barcinonensis OX=198119 GN=celA PE=1 SV=1 |
| P28621 | 2.04e-56 | 30 | 412 | 30 | 385 | Endoglucanase B OS=Clostridium cellulovorans (strain ATCC 35296 / DSM 3052 / OCM 3 / 743B) OX=573061 GN=engB PE=3 SV=1 |
| P17901 | 4.64e-56 | 39 | 399 | 44 | 398 | Endoglucanase A OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCA PE=1 SV=1 |
| P10477 | 9.81e-56 | 36 | 399 | 51 | 383 | Cellulase/esterase CelE OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celE PE=1 SV=2 |
| A7LXT7 | 7.47e-55 | 23 | 399 | 126 | 499 | Xyloglucan-specific endo-beta-1,4-glucanase BoGH5A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02653 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
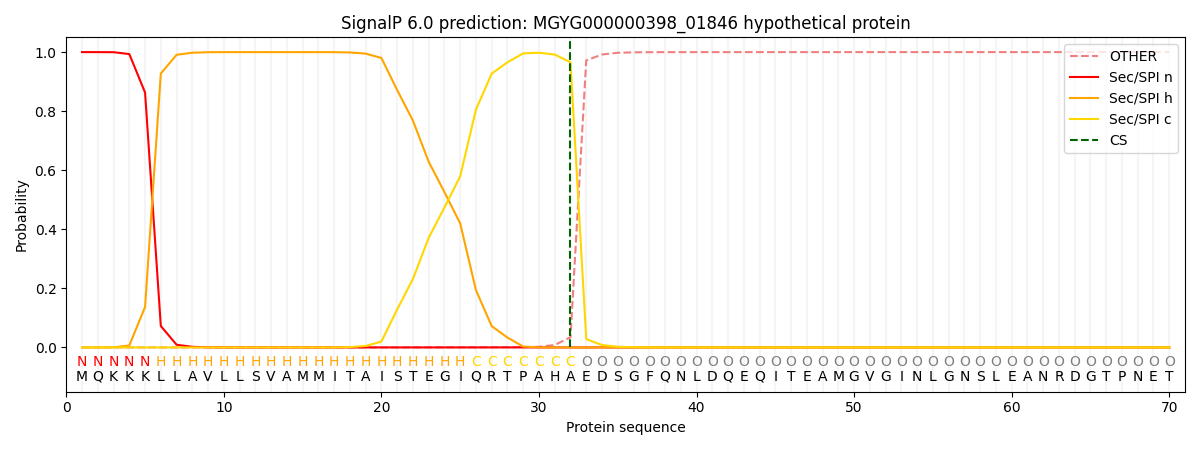
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000247 | 0.998784 | 0.000411 | 0.000188 | 0.000182 | 0.000154 |
