You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000399_00246
You are here: Home > Sequence: MGYG000000399_00246
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
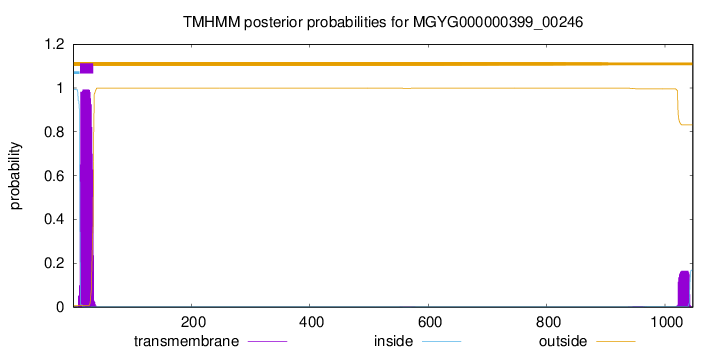
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; Borkfalkiaceae; ; | |||||||||||
| CAZyme ID | MGYG000000399_00246 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 51970; End: 55113 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 353 | 562 | 2.1e-19 | 0.5076923076923077 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 2.00e-25 | 299 | 533 | 83 | 323 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| PLN02218 | PLN02218 | 2.31e-08 | 298 | 522 | 67 | 267 | polygalacturonase ADPG |
| pfam12708 | Pectate_lyase_3 | 1.65e-04 | 300 | 337 | 3 | 41 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA23464.1 | 5.39e-121 | 55 | 783 | 84 | 799 |
| QCD41820.1 | 5.59e-82 | 81 | 727 | 239 | 878 |
| ADB48561.1 | 9.65e-39 | 289 | 871 | 20 | 573 |
| QGQ94767.1 | 3.78e-30 | 306 | 800 | 46 | 533 |
| ADB48560.1 | 1.81e-22 | 289 | 769 | 20 | 488 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3JUR_A | 1.14e-08 | 294 | 535 | 23 | 276 | Thecrystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_B The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_C The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima],3JUR_D The crystal structure of a hyperthermoactive Exopolygalacturonase from Thermotoga maritima [Thermotoga maritima] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8RY29 | 1.38e-07 | 298 | 522 | 67 | 267 | Polygalacturonase ADPG2 OS=Arabidopsis thaliana OX=3702 GN=ADPG2 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
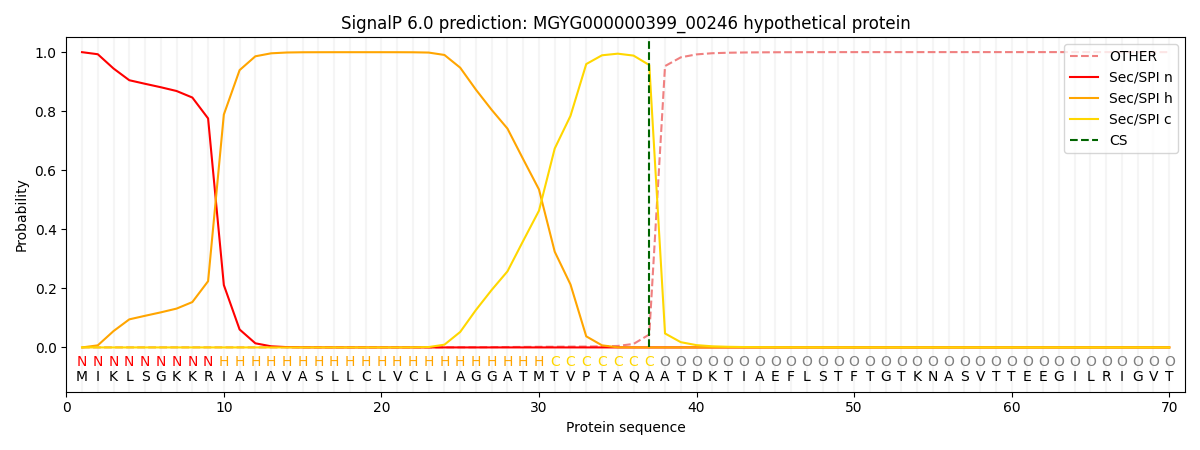
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000723 | 0.998425 | 0.000250 | 0.000228 | 0.000187 | 0.000165 |