You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000400_00949
You are here: Home > Sequence: MGYG000000400_00949
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; UBA7173; | |||||||||||
| CAZyme ID | MGYG000000400_00949 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 7621; End: 9210 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 64 | 415 | 7.8e-38 | 0.7262569832402235 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03663 | Glyco_hydro_76 | 3.04e-16 | 168 | 366 | 86 | 255 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| COG4833 | COG4833 | 6.06e-10 | 120 | 359 | 48 | 241 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT62205.1 | 3.27e-96 | 23 | 467 | 44 | 478 |
| QQA30660.1 | 3.27e-96 | 23 | 467 | 44 | 478 |
| QDH77939.1 | 1.62e-95 | 60 | 467 | 66 | 475 |
| SCD21417.1 | 1.66e-95 | 22 | 467 | 31 | 476 |
| AWW30266.1 | 2.28e-95 | 60 | 467 | 66 | 475 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MU9_A | 9.88e-16 | 74 | 400 | 53 | 300 | Crystalstructure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482],4MU9_B Crystal structure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
| 6SHD_A | 1.64e-13 | 166 | 354 | 131 | 281 | Structureof the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_B Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_C Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6] |
| 4C1S_A | 6.34e-13 | 168 | 468 | 107 | 369 | Glycosidehydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],4C1S_B Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 6Y8F_A | 1.23e-12 | 166 | 354 | 132 | 282 | ChainA, Alpha-1,6-endo-mannanase GH76A mutant [Salegentibacter sp. Hel_I_6] |
| 6SHM_A | 3.88e-12 | 173 | 354 | 140 | 282 | Aninactive (D136A and D137A) variant of alpha-1,6-mannanase, GH76A of Salegentibacter sp. HEL1_6 in complex with alpha-1,6-mannotetrose [Salegentibacter sp. Hel_I_6] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
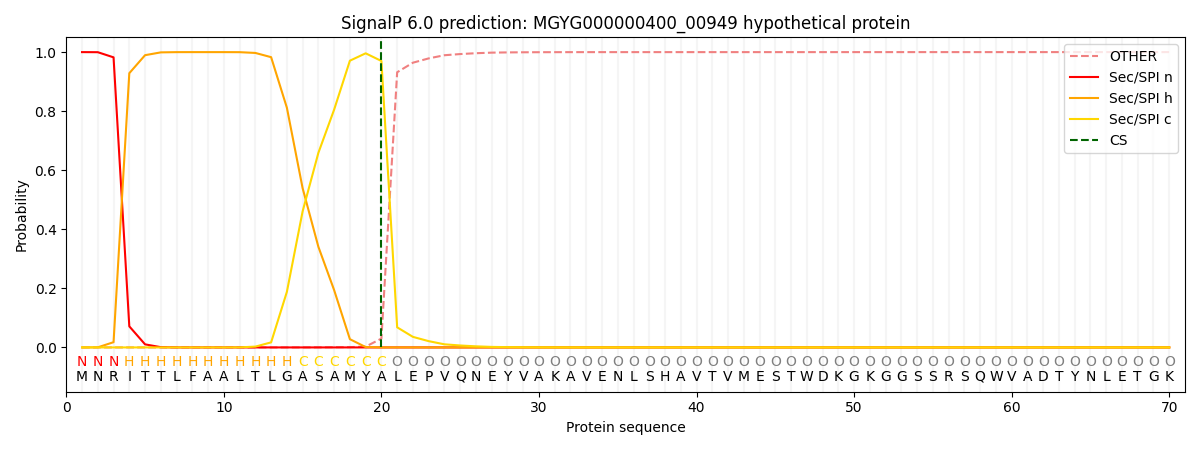
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000294 | 0.999008 | 0.000168 | 0.000172 | 0.000161 | 0.000154 |
