You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000414_00782
You are here: Home > Sequence: MGYG000000414_00782
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes sp900546065 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes sp900546065 | |||||||||||
| CAZyme ID | MGYG000000414_00782 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 390527; End: 391639 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 50 | 334 | 1.6e-116 | 0.9930313588850174 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG2730 | BglC | 1.87e-28 | 45 | 364 | 49 | 392 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam00150 | Cellulase | 6.77e-20 | 59 | 334 | 21 | 268 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| SCM55578.1 | 1.06e-159 | 48 | 368 | 44 | 365 |
| ATP56029.1 | 7.18e-123 | 47 | 369 | 37 | 353 |
| ARA93352.1 | 6.49e-120 | 44 | 369 | 26 | 348 |
| AQG82496.1 | 6.63e-110 | 48 | 369 | 38 | 360 |
| QJW89098.1 | 7.56e-110 | 48 | 369 | 42 | 364 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7EC9_A | 5.70e-89 | 48 | 369 | 20 | 341 | ChainA, Endoglucanase [Thermotoga maritima MSB8],7EC9_B Chain B, Endoglucanase [Thermotoga maritima MSB8],7EFZ_A Chain A, Endoglucanase [Thermotoga maritima MSB8],7EFZ_B Chain B, Endoglucanase [Thermotoga maritima MSB8] |
| 1VJZ_A | 1.46e-86 | 48 | 369 | 20 | 341 | Crystalstructure of Endoglucanase (TM1752) from Thermotoga maritima at 2.05 A resolution [Thermotoga maritima] |
| 3W0K_A | 2.72e-78 | 71 | 367 | 29 | 326 | CrystalStructure of a glycoside hydrolase [Caldanaerobius polysaccharolyticus],3W0K_B Crystal Structure of a glycoside hydrolase [Caldanaerobius polysaccharolyticus] |
| 3AMC_A | 1.03e-21 | 45 | 345 | 10 | 299 | Crystalstructures of Thermotoga maritima Cel5A, apo form and dimer/au [Thermotoga maritima MSB8],3AMC_B Crystal structures of Thermotoga maritima Cel5A, apo form and dimer/au [Thermotoga maritima MSB8],3AMD_A Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_B Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_C Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3AMD_D Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au [Thermotoga maritima MSB8],3MMU_A Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_B Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_C Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_D Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_E Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_F Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_G Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMU_H Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_A Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_B Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_C Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima],3MMW_D Crystal structure of endoglucanase Cel5A from the hyperthermophilic Thermotoga maritima [Thermotoga maritima] |
| 3AMG_A | 3.63e-21 | 45 | 345 | 10 | 299 | Crystalstructures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form [Thermotoga maritima MSB8],3AMG_B Crystal structures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form [Thermotoga maritima MSB8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16169 | 5.76e-19 | 67 | 326 | 33 | 285 | Cellodextrinase A OS=Ruminococcus flavefaciens OX=1265 GN=celA PE=3 SV=3 |
| Q5AVZ7 | 1.51e-07 | 50 | 130 | 461 | 541 | Glucan 1,3-beta-glucosidase D OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=exgD PE=2 SV=1 |
| P32603 | 3.45e-07 | 63 | 191 | 113 | 248 | Sporulation-specific glucan 1,3-beta-glucosidase OS=Saccharomyces cerevisiae (strain ATCC 204508 / S288c) OX=559292 GN=SPR1 PE=2 SV=1 |
| Q12628 | 7.89e-07 | 55 | 225 | 87 | 223 | Glucan 1,3-beta-glucosidase OS=Kluyveromyces lactis (strain ATCC 8585 / CBS 2359 / DSM 70799 / NBRC 1267 / NRRL Y-1140 / WM37) OX=284590 GN=KLLA0C05324g PE=3 SV=1 |
| Q876J2 | 1.08e-06 | 63 | 178 | 113 | 233 | Sporulation-specific glucan 1,3-beta-glucosidase OS=Saccharomyces uvarum (strain ATCC 76518 / CBS 7001 / CLIB 283 / NBRC 10550 / MCYC 623 / NCYC 2669 / NRRL Y-11845) OX=659244 GN=SPR1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as TAT
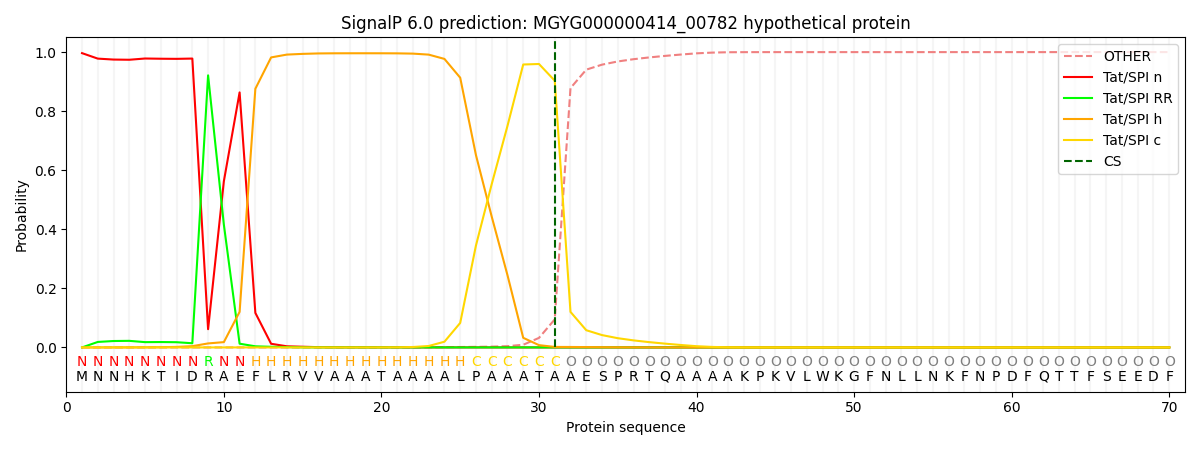
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000010 | 0.000001 | 0.996500 | 0.003465 | 0.000000 |

