You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000414_01031
You are here: Home > Sequence: MGYG000000414_01031
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes sp900546065 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes sp900546065 | |||||||||||
| CAZyme ID | MGYG000000414_01031 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 139497; End: 140516 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 90 | 305 | 8.9e-29 | 0.8339100346020761 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 6.89e-12 | 93 | 295 | 66 | 265 | Cellulase (glycosyl hydrolase family 5). |
| COG3934 | COG3934 | 3.15e-07 | 93 | 322 | 68 | 288 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| COG2730 | BglC | 3.23e-06 | 101 | 251 | 125 | 273 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| pfam02836 | Glyco_hydro_2_C | 0.001 | 137 | 274 | 97 | 217 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| COG3250 | LacZ | 0.006 | 136 | 206 | 367 | 438 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ATB28678.1 | 2.19e-108 | 29 | 339 | 46 | 356 |
| QRN97019.1 | 2.83e-100 | 29 | 334 | 45 | 350 |
| ATB43865.1 | 5.75e-100 | 29 | 332 | 45 | 348 |
| AKJ31985.1 | 1.47e-69 | 17 | 330 | 2 | 317 |
| AGZ41046.1 | 2.45e-65 | 16 | 312 | 32 | 319 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3WFL_A | 4.33e-13 | 93 | 275 | 90 | 284 | Crtstalstructure of glycoside hydrolase family 5 beta-mannanase from Talaromyces trachyspermus [Talaromyces trachyspermus] |
| 4QP0_A | 4.12e-10 | 29 | 279 | 4 | 305 | CrystalStructure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
| 1RH9_A | 1.85e-09 | 29 | 268 | 6 | 294 | ChainA, endo-beta-mannanase [Solanum lycopersicum] |
| 3ZIZ_A | 4.52e-09 | 93 | 279 | 102 | 298 | ChainA, Gh5 Endo-beta-1,4-mannanase [Podospora anserina] |
| 3PZ9_A | 1.44e-08 | 93 | 330 | 101 | 379 | Nativestructure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0IQJ7 | 7.15e-15 | 33 | 309 | 5 | 340 | Mannan endo-1,4-beta-mannosidase 8 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN8 PE=2 SV=2 |
| Q2U2I3 | 1.89e-14 | 93 | 312 | 208 | 432 | Probable mannan endo-1,4-beta-mannosidase F OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=manF PE=3 SV=2 |
| B8NIV9 | 1.89e-14 | 93 | 312 | 208 | 432 | Probable mannan endo-1,4-beta-mannosidase F OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=manF PE=3 SV=1 |
| Q9SG94 | 2.92e-14 | 8 | 339 | 7 | 395 | Mannan endo-1,4-beta-mannosidase 3 OS=Arabidopsis thaliana OX=3702 GN=MAN3 PE=2 SV=1 |
| Q2RBB1 | 6.06e-14 | 32 | 309 | 14 | 347 | Mannan endo-1,4-beta-mannosidase 7 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN7 PE=2 SV=3 |
SignalP and Lipop Annotations help
This protein is predicted as SP
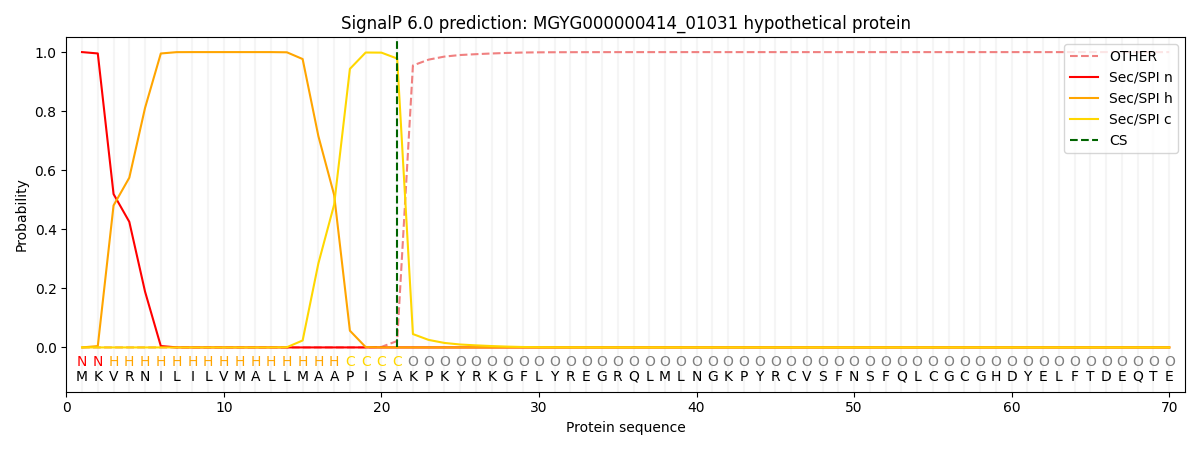
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000244 | 0.999180 | 0.000165 | 0.000137 | 0.000138 | 0.000131 |
