You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000415_00351
You are here: Home > Sequence: MGYG000000415_00351
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
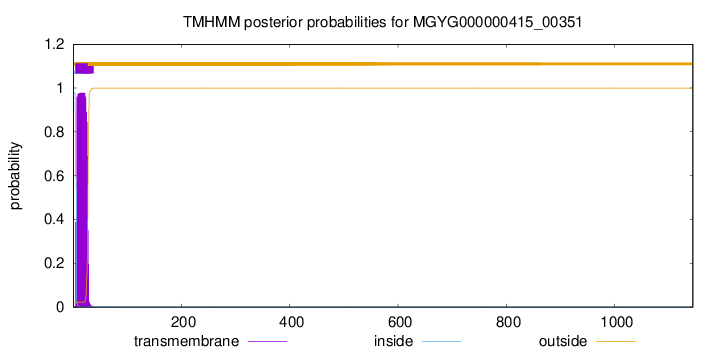
TMHMM annotations
Basic Information help
| Species | UBA11471 sp900547555 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA11471; UBA11471; UBA11471 sp900547555 | |||||||||||
| CAZyme ID | MGYG000000415_00351 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 379314; End: 382748 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 735 | 1015 | 1.8e-51 | 0.9513888888888888 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 3.31e-24 | 731 | 1018 | 89 | 404 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| pfam01095 | Pectinesterase | 1.68e-21 | 737 | 1018 | 16 | 297 | Pectinesterase. |
| PLN02665 | PLN02665 | 5.76e-19 | 828 | 1023 | 175 | 360 | pectinesterase family protein |
| PLN02708 | PLN02708 | 1.50e-17 | 736 | 1025 | 253 | 551 | Probable pectinesterase/pectinesterase inhibitor |
| PLN02488 | PLN02488 | 1.95e-17 | 718 | 1025 | 194 | 500 | probable pectinesterase/pectinesterase inhibitor |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ASB37302.1 | 2.05e-264 | 2 | 1133 | 4 | 1141 |
| ANU64595.1 | 2.05e-264 | 2 | 1133 | 4 | 1141 |
| QQR08039.1 | 2.05e-264 | 2 | 1133 | 4 | 1141 |
| QUT74167.1 | 1.94e-84 | 593 | 1131 | 882 | 1414 |
| QCD38476.1 | 5.89e-75 | 659 | 1131 | 972 | 1455 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q3EAY9 | 1.55e-12 | 754 | 982 | 216 | 453 | Probable pectinesterase 30 OS=Arabidopsis thaliana OX=3702 GN=PME30 PE=2 SV=1 |
| Q1PEC0 | 5.11e-12 | 718 | 1023 | 211 | 513 | Probable pectinesterase/pectinesterase inhibitor 42 OS=Arabidopsis thaliana OX=3702 GN=PME42 PE=2 SV=1 |
| Q84JX1 | 8.18e-11 | 718 | 1023 | 211 | 513 | Probable pectinesterase/pectinesterase inhibitor 19 OS=Arabidopsis thaliana OX=3702 GN=PME19 PE=2 SV=1 |
| O49298 | 6.01e-10 | 754 | 1018 | 280 | 539 | Probable pectinesterase/pectinesterase inhibitor 6 OS=Arabidopsis thaliana OX=3702 GN=PME6 PE=2 SV=1 |
| P17872 | 3.21e-08 | 732 | 973 | 36 | 289 | Pectinesterase OS=Aspergillus niger OX=5061 GN=pme1 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
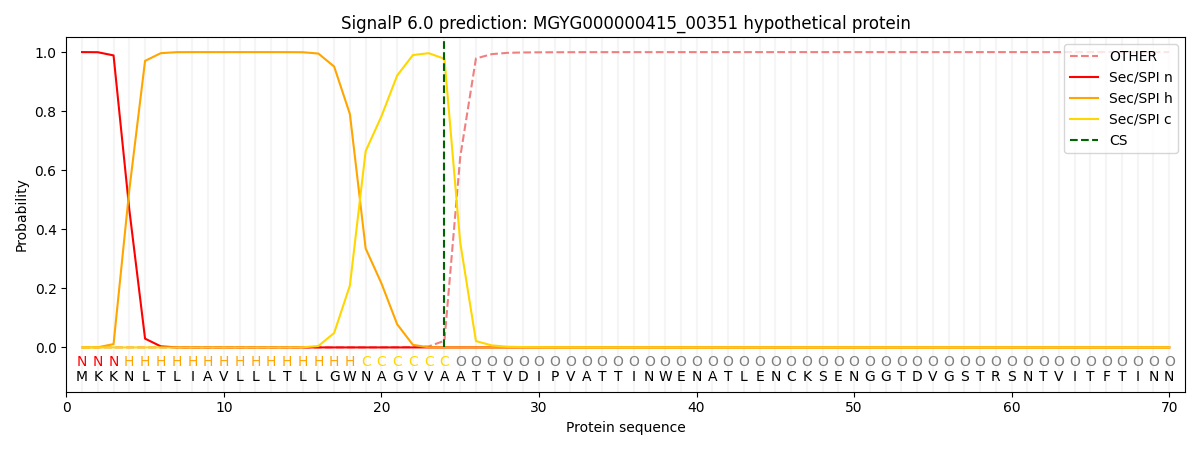
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000349 | 0.998951 | 0.000169 | 0.000193 | 0.000161 | 0.000158 |