You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000415_00371
You are here: Home > Sequence: MGYG000000415_00371
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA11471 sp900547555 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA11471; UBA11471; UBA11471 sp900547555 | |||||||||||
| CAZyme ID | MGYG000000415_00371 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 400906; End: 404019 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 501 | 687 | 9.4e-54 | 0.994535519125683 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| TIGR04183 | Por_Secre_tail | 1.70e-04 | 969 | 1035 | 9 | 71 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
| NF033708 | T9SS_Cterm_ChiA | 0.006 | 980 | 1034 | 1 | 54 | T9SS sorting signal type C. The sorting signals of type IX secretion systems (T9SS) in the CFB bacteria are long, compared to other prokaryotic C-terminal sorting motif-containing signals, including LPXTG, PEP-CTERM, and GlyGly-TERM, and they seem to contain multiple motifs. A few T9SS substrates, including ChiA, have a variant form of T9SS sorting signal that may score poorly to both TIGR04183 (type A) and TIGR04131 (type B), depend on T9SS for secretion, but are released from the cell rather than left anchored to the cell surface. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT77843.1 | 3.50e-202 | 342 | 1035 | 206 | 923 |
| AWW33033.1 | 3.28e-183 | 292 | 964 | 159 | 826 |
| QDH81626.1 | 6.51e-183 | 292 | 964 | 159 | 826 |
| AWK06451.1 | 1.01e-182 | 283 | 962 | 141 | 818 |
| QOG03624.1 | 7.90e-182 | 342 | 962 | 202 | 818 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NQQ7 | 9.80e-40 | 440 | 857 | 12 | 393 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| Q0CLG7 | 9.80e-40 | 448 | 857 | 20 | 393 | Probable pectate lyase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyC PE=3 SV=1 |
| Q2UB83 | 2.42e-39 | 440 | 857 | 12 | 393 | Probable pectate lyase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyC PE=3 SV=1 |
| Q5B297 | 2.52e-39 | 448 | 877 | 20 | 404 | Probable pectate lyase C OS=Emericella nidulans (strain FGSC A4 / ATCC 38163 / CBS 112.46 / NRRL 194 / M139) OX=227321 GN=plyC PE=3 SV=1 |
| A1DPF0 | 3.39e-36 | 448 | 856 | 21 | 393 | Probable pectate lyase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
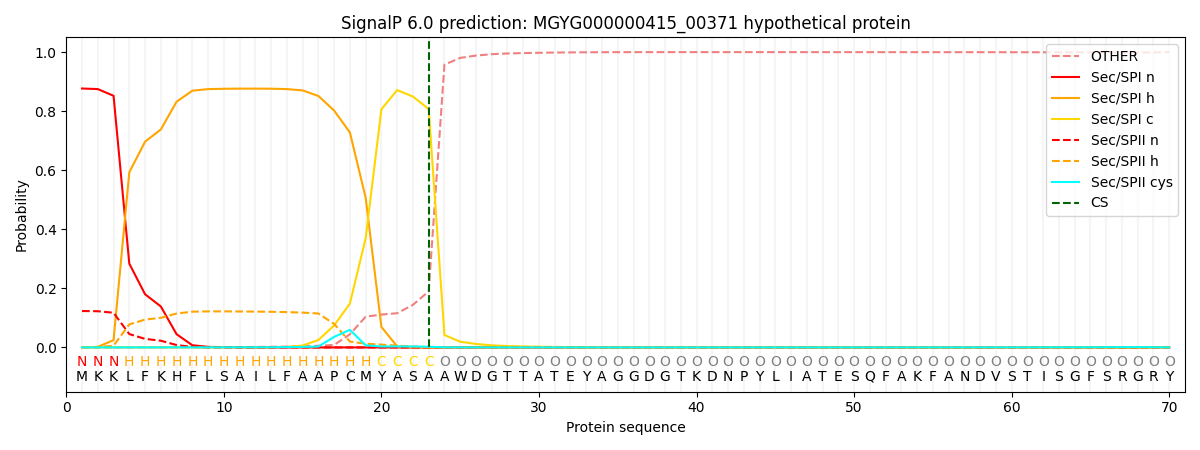
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000738 | 0.868364 | 0.130014 | 0.000328 | 0.000289 | 0.000259 |
