You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000424_00872
You are here: Home > Sequence: MGYG000000424_00872
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-353 sp900768995 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; CAG-353; CAG-353 sp900768995 | |||||||||||
| CAZyme ID | MGYG000000424_00872 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 25881; End: 27017 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 62 | 348 | 3.3e-55 | 0.9868852459016394 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG2730 | BglC | 0.004 | 49 | 210 | 28 | 193 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| smart00636 | Glyco_18 | 0.006 | 115 | 194 | 34 | 107 | Glyco_18 domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL16471.1 | 2.30e-141 | 48 | 376 | 31 | 361 |
| CCO03823.1 | 4.36e-133 | 3 | 376 | 4 | 396 |
| ADD61974.1 | 3.36e-132 | 50 | 376 | 71 | 405 |
| QHW32543.1 | 4.97e-117 | 50 | 376 | 36 | 366 |
| QHT63496.1 | 7.24e-117 | 50 | 376 | 36 | 366 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5Y6T_A | 4.07e-28 | 43 | 349 | 1 | 330 | ChainA, endo-1,4-beta-mannanase [Eisenia fetida] |
| 4OOU_A | 7.81e-22 | 45 | 353 | 18 | 349 | Crystalstructure of beta-1,4-D-mannanase from Cryptopygus antarcticus [Cryptopygus antarcticus],4OOU_B Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus [Cryptopygus antarcticus],4OOZ_A Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus in complex with mannopentaose [Cryptopygus antarcticus],4OOZ_B Crystal structure of beta-1,4-D-mannanase from Cryptopygus antarcticus in complex with mannopentaose [Cryptopygus antarcticus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B4XC07 | 4.02e-21 | 45 | 353 | 18 | 349 | Mannan endo-1,4-beta-mannosidase OS=Cryptopygus antarcticus OX=187623 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
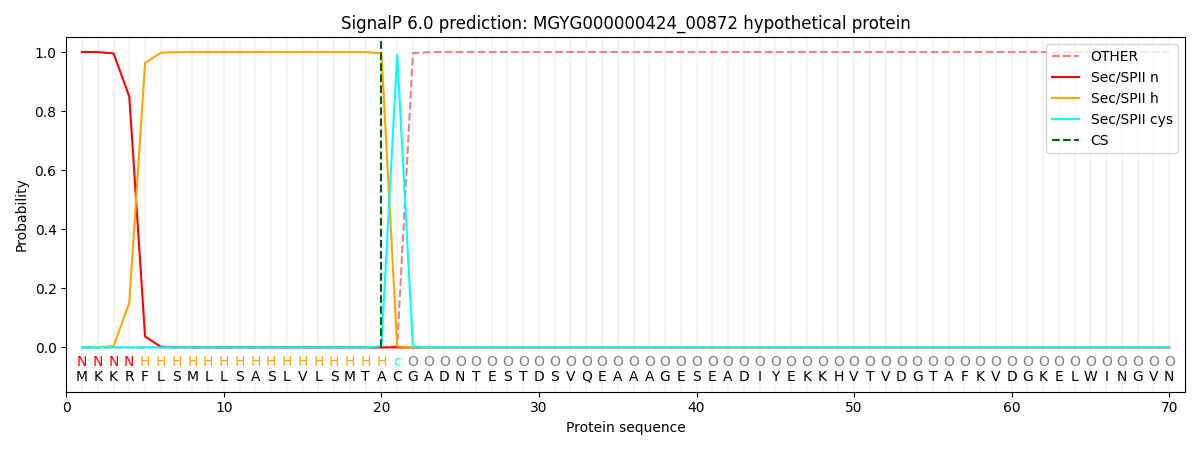
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000045 | 0.000000 | 0.000000 | 0.000000 |
