You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000435_01069
You are here: Home > Sequence: MGYG000000435_01069
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RUG131 sp900549975 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; RFN20; CAG-826; RUG131; RUG131 sp900549975 | |||||||||||
| CAZyme ID | MGYG000000435_01069 | |||||||||||
| CAZy Family | GH138 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 93966; End: 96161 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH138 | 4 | 695 | 3.2e-70 | 0.7442116868798236 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02838 | Glyco_hydro_20b | 2.69e-07 | 21 | 100 | 21 | 100 | Glycosyl hydrolase family 20, domain 2. This domain has a zincin-like fold. |
| COG0266 | Nei | 2.41e-06 | 607 | 651 | 130 | 177 | Formamidopyrimidine-DNA glycosylase [Replication, recombination and repair]. |
| PRK01103 | PRK01103 | 2.56e-04 | 595 | 651 | 118 | 177 | bifunctional DNA-formamidopyrimidine glycosylase/DNA-(apurinic or apyrimidinic site) lyase. |
| pfam06831 | H2TH | 2.80e-04 | 617 | 651 | 11 | 45 | Formamidopyrimidine-DNA glycosylase H2TH domain. Formamidopyrimidine-DNA glycosylase (Fpg) is a DNA repair enzyme that excises oxidized purines from damaged DNA. This family is the central domain containing the DNA-binding helix-two turn-helix domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ARV16526.1 | 4.72e-113 | 1 | 727 | 1 | 720 |
| QCW99550.1 | 1.30e-112 | 12 | 723 | 15 | 723 |
| QDV30222.1 | 3.11e-108 | 13 | 727 | 29 | 733 |
| QDV30507.1 | 8.49e-102 | 32 | 727 | 32 | 720 |
| AUP80499.1 | 1.37e-101 | 19 | 727 | 2 | 706 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
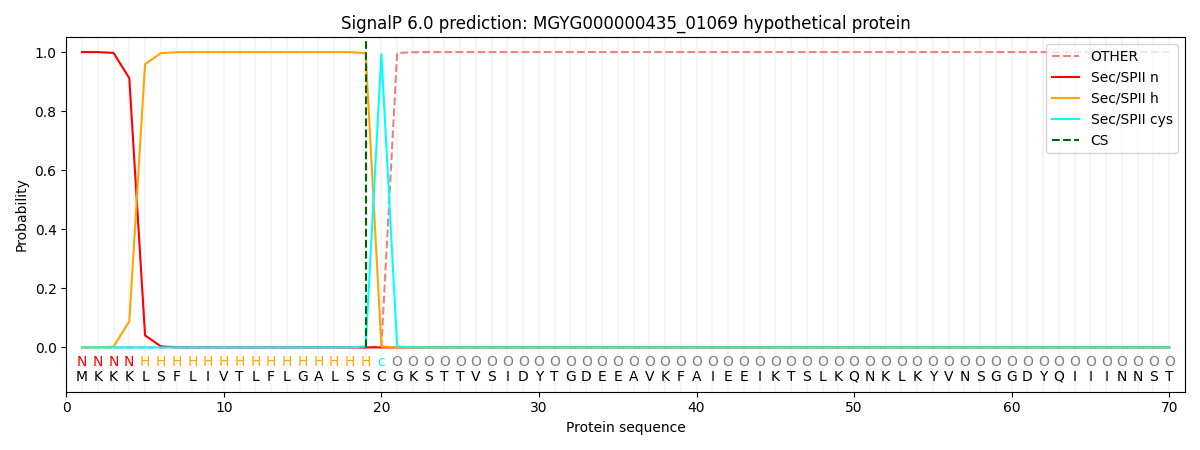
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000054 | 0.000000 | 0.000000 | 0.000000 |
