You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000437_00485
You are here: Home > Sequence: MGYG000000437_00485
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes sp001941065 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes sp001941065 | |||||||||||
| CAZyme ID | MGYG000000437_00485 | |||||||||||
| CAZy Family | GH33 | |||||||||||
| CAZyme Description | Sialidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 21046; End: 22200 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH33 | 38 | 378 | 1.7e-90 | 0.9678362573099415 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd15482 | Sialidase_non-viral | 3.92e-100 | 38 | 381 | 1 | 339 | Non-viral sialidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates, they play vital roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed, beta-propeller fold with the non-viral sialidases containing 2-5 Asp-box motifs (most commonly Ser/Thr-X-Asp-[X]-Gly-X-Thr- Trp/Phe). This CD includes eubacterial and eukaryotic sialidases. |
| pfam13088 | BNR_2 | 3.72e-33 | 62 | 361 | 1 | 280 | BNR repeat-like domain. This family of proteins contains BNR-like repeats suggesting these proteins may act as sialidases. |
| COG4409 | NanH | 2.08e-08 | 60 | 364 | 376 | 700 | Neuraminidase (sialidase) [Carbohydrate transport and metabolism, Cell wall/membrane/envelope biogenesis]. |
| cd00260 | Sialidase | 6.94e-05 | 51 | 183 | 15 | 143 | sialidases/neuraminidases. Sialidases or neuraminidases function to bind and hydrolyze terminal sialic acid residues from various glycoconjugates as well as playing roles in pathogenesis, bacterial nutrition and cellular interactions. They have a six-bladed beta-propeller fold. This hierarchy includes eubacterial, eukaryotic, and viral sialidases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BBD46002.1 | 4.76e-107 | 37 | 377 | 36 | 367 |
| QDV16491.1 | 3.02e-102 | 39 | 383 | 40 | 379 |
| QDT26944.1 | 6.05e-102 | 39 | 383 | 40 | 379 |
| QDU50208.1 | 6.05e-102 | 39 | 383 | 40 | 379 |
| QDT20890.1 | 1.71e-101 | 39 | 383 | 40 | 379 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4FJ6_A | 6.82e-37 | 44 | 376 | 174 | 516 | Crystalstructure of a glycoside hydrolase family 33, candidate sialidase (BDI_2946) from Parabacteroides distasonis ATCC 8503 at 1.90 A resolution [Parabacteroides distasonis ATCC 8503],4FJ6_B Crystal structure of a glycoside hydrolase family 33, candidate sialidase (BDI_2946) from Parabacteroides distasonis ATCC 8503 at 1.90 A resolution [Parabacteroides distasonis ATCC 8503],4FJ6_C Crystal structure of a glycoside hydrolase family 33, candidate sialidase (BDI_2946) from Parabacteroides distasonis ATCC 8503 at 1.90 A resolution [Parabacteroides distasonis ATCC 8503],4FJ6_D Crystal structure of a glycoside hydrolase family 33, candidate sialidase (BDI_2946) from Parabacteroides distasonis ATCC 8503 at 1.90 A resolution [Parabacteroides distasonis ATCC 8503] |
| 6MNJ_A | 3.24e-36 | 44 | 368 | 193 | 525 | Hadzamicrobial sialidase Hz136 [Alistipes],6MNJ_B Hadza microbial sialidase Hz136 [Alistipes] |
| 6MYV_A | 6.61e-36 | 44 | 383 | 174 | 518 | Sialidase26co-crystallized with DANA-Gc [bacterium],6MYV_B Sialidase26 co-crystallized with DANA-Gc [bacterium],6MYV_C Sialidase26 co-crystallized with DANA-Gc [bacterium],6MYV_D Sialidase26 co-crystallized with DANA-Gc [bacterium] |
| 6MRV_A | 8.74e-36 | 44 | 383 | 195 | 539 | Sialidase26co-crystallized with DANA [bacterium],6MRV_B Sialidase26 co-crystallized with DANA [bacterium],6MRX_A Sialidase26 apo [bacterium],6MRX_B Sialidase26 apo [bacterium],6MRX_C Sialidase26 apo [bacterium],6MRX_D Sialidase26 apo [bacterium] |
| 4Q6K_A | 1.57e-34 | 44 | 375 | 177 | 518 | Crystalstructure of a putative neuraminidase (BACCAC_01090) from Bacteroides caccae ATCC 43185 at 1.90 A resolution (PSI Community Target) [Bacteroides caccae ATCC 43185],4Q6K_B Crystal structure of a putative neuraminidase (BACCAC_01090) from Bacteroides caccae ATCC 43185 at 1.90 A resolution (PSI Community Target) [Bacteroides caccae ATCC 43185],4Q6K_C Crystal structure of a putative neuraminidase (BACCAC_01090) from Bacteroides caccae ATCC 43185 at 1.90 A resolution (PSI Community Target) [Bacteroides caccae ATCC 43185],4Q6K_D Crystal structure of a putative neuraminidase (BACCAC_01090) from Bacteroides caccae ATCC 43185 at 1.90 A resolution (PSI Community Target) [Bacteroides caccae ATCC 43185] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A6BMK7 | 1.30e-31 | 51 | 366 | 77 | 396 | Sialidase-1 OS=Bos taurus OX=9913 GN=NEU1 PE=2 SV=2 |
| A5PF10 | 1.32e-31 | 51 | 366 | 78 | 397 | Sialidase-1 OS=Sus scrofa OX=9823 GN=NEU1 PE=3 SV=1 |
| O35657 | 3.13e-31 | 51 | 366 | 71 | 390 | Sialidase-1 OS=Mus musculus OX=10090 GN=Neu1 PE=1 SV=1 |
| Q99519 | 4.71e-31 | 51 | 366 | 77 | 396 | Sialidase-1 OS=Homo sapiens OX=9606 GN=NEU1 PE=1 SV=1 |
| Q5RAF4 | 4.71e-31 | 51 | 366 | 77 | 396 | Sialidase-1 OS=Pongo abelii OX=9601 GN=NEU1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
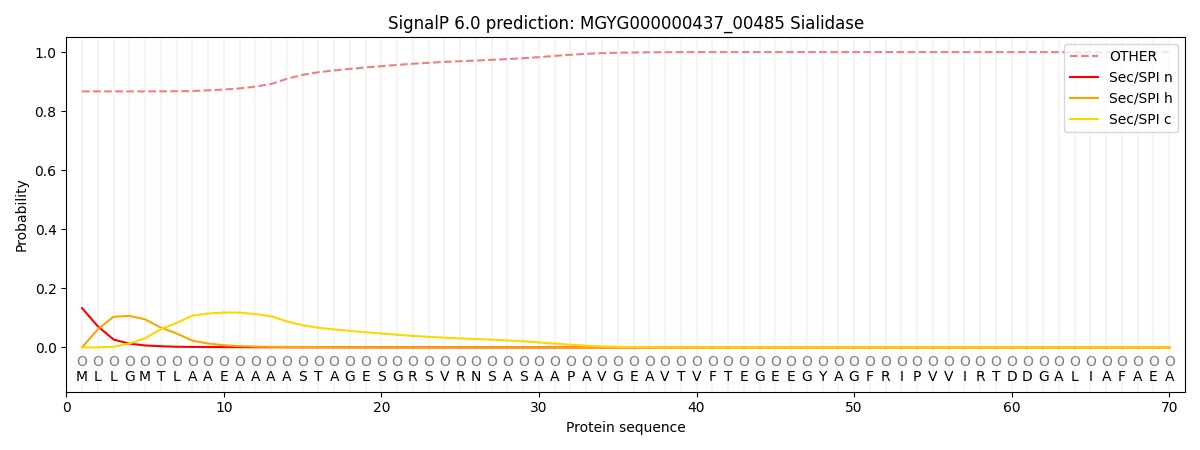
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.873199 | 0.126323 | 0.000150 | 0.000154 | 0.000088 | 0.000105 |
