You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000437_00718
You are here: Home > Sequence: MGYG000000437_00718
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Alistipes sp001941065 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Rikenellaceae; Alistipes; Alistipes sp001941065 | |||||||||||
| CAZyme ID | MGYG000000437_00718 | |||||||||||
| CAZy Family | GH49 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12733; End: 14298 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH49 | 177 | 509 | 8.5e-41 | 0.6575591985428051 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ANH83078.1 | 7.03e-213 | 51 | 521 | 48 | 520 |
| BBL03438.1 | 1.69e-180 | 46 | 521 | 52 | 526 |
| EDY96541.1 | 1.24e-128 | 52 | 516 | 17 | 473 |
| QCT01546.1 | 3.66e-49 | 112 | 511 | 117 | 537 |
| QBN20516.1 | 2.37e-48 | 41 | 503 | 16 | 444 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6NZS_A | 1.59e-24 | 52 | 504 | 25 | 564 | DextranaseAoDex KQ11 [Pseudarthrobacter oxydans] |
| 1WMR_A | 1.28e-15 | 143 | 511 | 134 | 535 | CrystalStructure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],1WMR_B Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],1X0C_A Improved Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],1X0C_B Improved Crystal Structure of Isopullulanase from Aspergillus niger ATCC 9642 [Aspergillus niger],2Z8G_A Aspergillus niger ATCC9642 isopullulanase complexed with isopanose [Aspergillus niger],2Z8G_B Aspergillus niger ATCC9642 isopullulanase complexed with isopanose [Aspergillus niger] |
| 3WWG_A | 1.28e-15 | 143 | 511 | 134 | 535 | Crystalstructure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger],3WWG_B Crystal structure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger],3WWG_C Crystal structure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger],3WWG_D Crystal structure of the N-glycan-deficient variant N448A of isopullulanase complexed with isopanose [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P70744 | 4.31e-24 | 58 | 512 | 82 | 603 | Dextranase OS=Arthrobacter globiformis OX=1665 PE=3 SV=1 |
| P39652 | 1.03e-23 | 58 | 443 | 82 | 542 | Dextranase OS=Arthrobacter sp. (strain CB-8) OX=74565 PE=1 SV=1 |
| O00105 | 7.27e-15 | 143 | 511 | 149 | 550 | Isopullulanase OS=Aspergillus niger OX=5061 GN=ipuA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
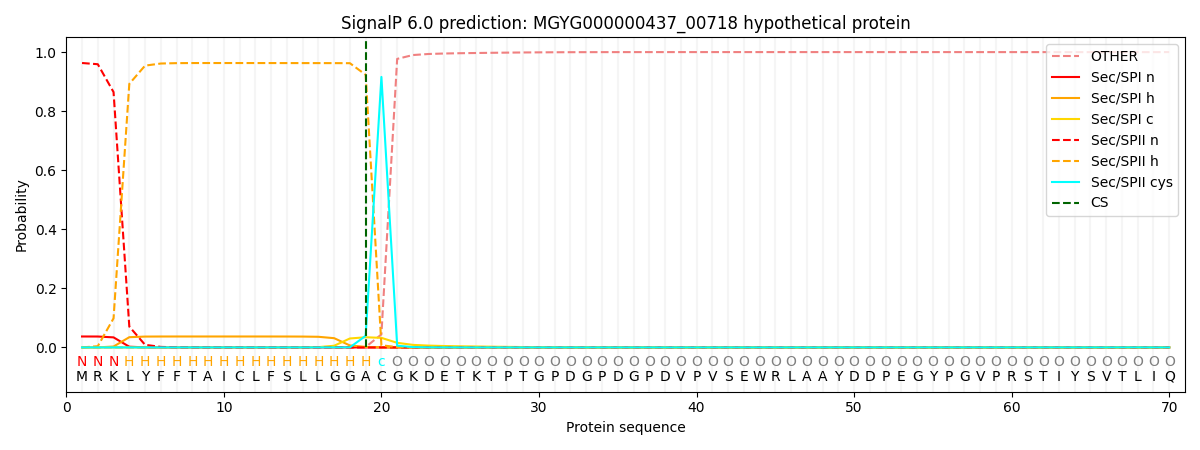
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000074 | 0.036086 | 0.963819 | 0.000015 | 0.000020 | 0.000017 |
